Calcium in PDB 8qzc: Structure of Calcium-Bound MTMEM16A(Ac)-L647V/I733V Chloride Channel at 3.29 A Resolution
Calcium Binding Sites:
The binding sites of Calcium atom in the Structure of Calcium-Bound MTMEM16A(Ac)-L647V/I733V Chloride Channel at 3.29 A Resolution
(pdb code 8qzc). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 4 binding sites of Calcium where determined in the Structure of Calcium-Bound MTMEM16A(Ac)-L647V/I733V Chloride Channel at 3.29 A Resolution, PDB code: 8qzc:
Jump to Calcium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Calcium where determined in the Structure of Calcium-Bound MTMEM16A(Ac)-L647V/I733V Chloride Channel at 3.29 A Resolution, PDB code: 8qzc:
Jump to Calcium binding site number: 1; 2; 3; 4;
Calcium binding site 1 out of 4 in 8qzc
Go back to
Calcium binding site 1 out
of 4 in the Structure of Calcium-Bound MTMEM16A(Ac)-L647V/I733V Chloride Channel at 3.29 A Resolution

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Structure of Calcium-Bound MTMEM16A(Ac)-L647V/I733V Chloride Channel at 3.29 A Resolution within 5.0Å range:
|
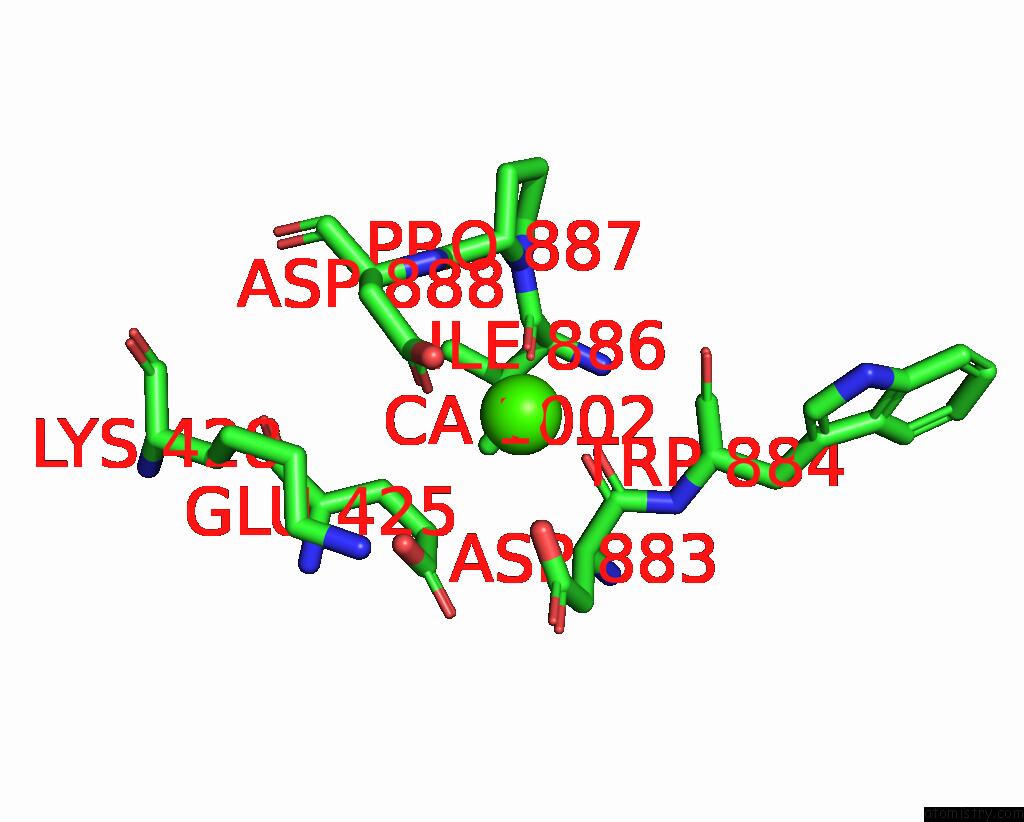
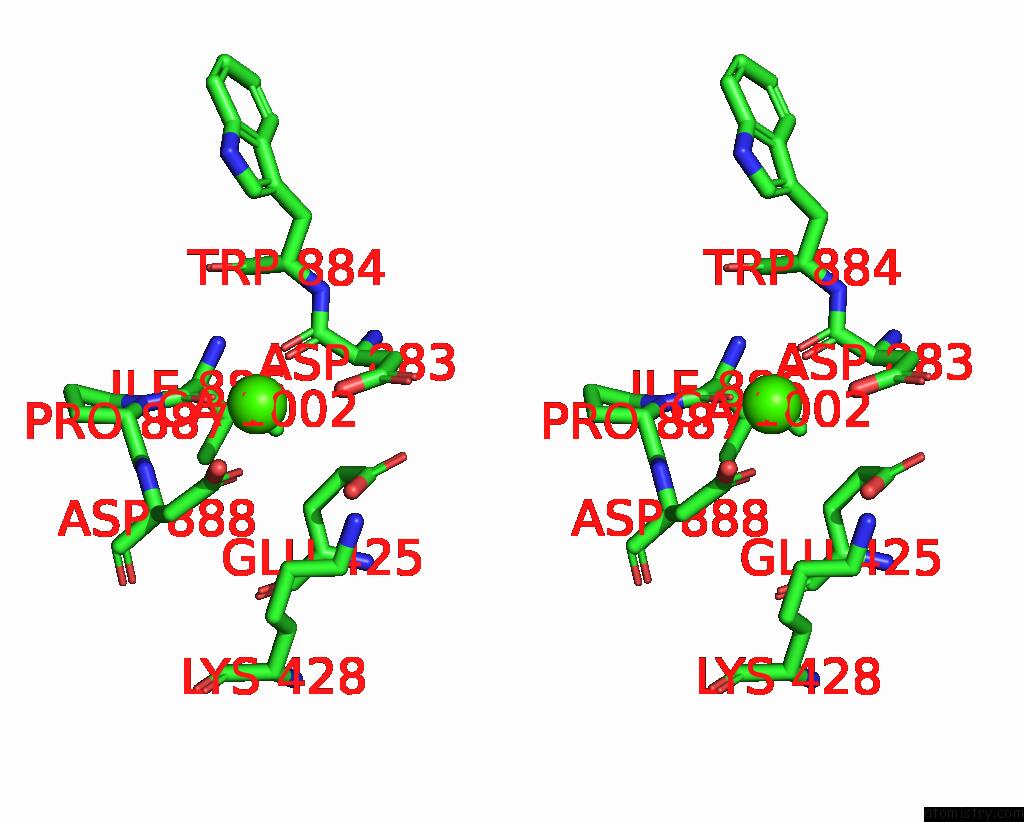
Calcium binding site 2 out of 4 in 8qzc
Go back to
Calcium binding site 2 out
of 4 in the Structure of Calcium-Bound MTMEM16A(Ac)-L647V/I733V Chloride Channel at 3.29 A Resolution
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Structure of Calcium-Bound MTMEM16A(Ac)-L647V/I733V Chloride Channel at 3.29 A Resolution within 5.0Å range:
|
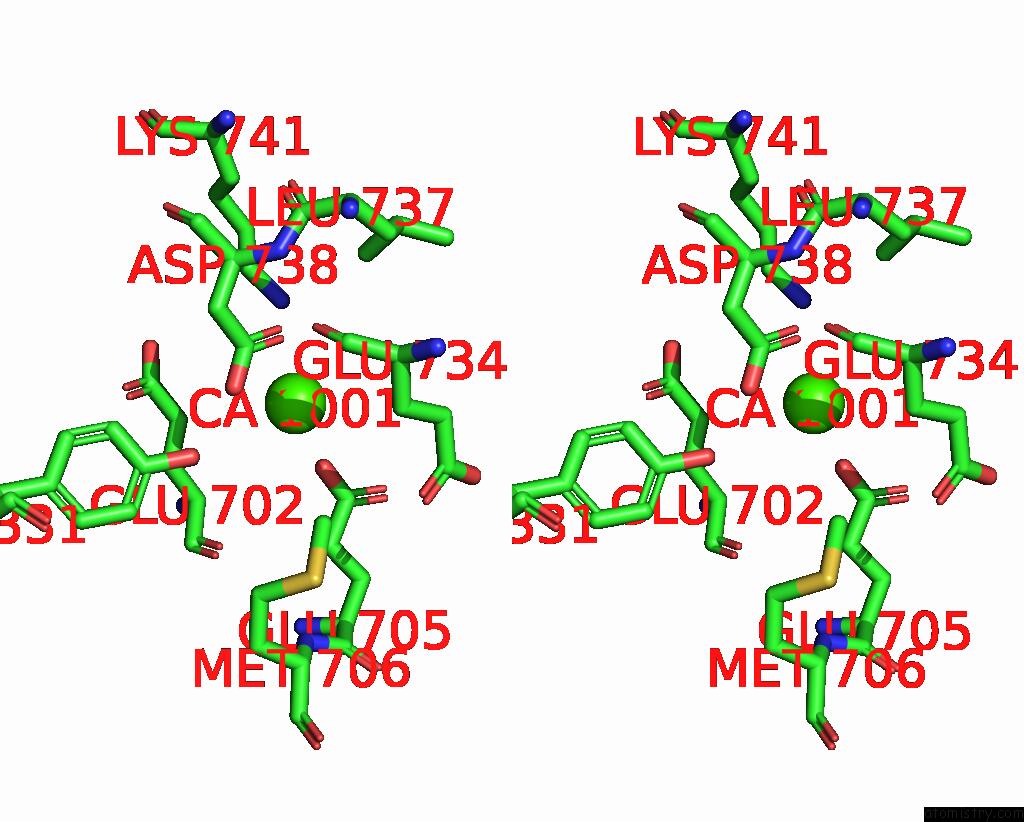
Calcium binding site 3 out of 4 in 8qzc
Go back to
Calcium binding site 3 out
of 4 in the Structure of Calcium-Bound MTMEM16A(Ac)-L647V/I733V Chloride Channel at 3.29 A Resolution

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Structure of Calcium-Bound MTMEM16A(Ac)-L647V/I733V Chloride Channel at 3.29 A Resolution within 5.0Å range:
|
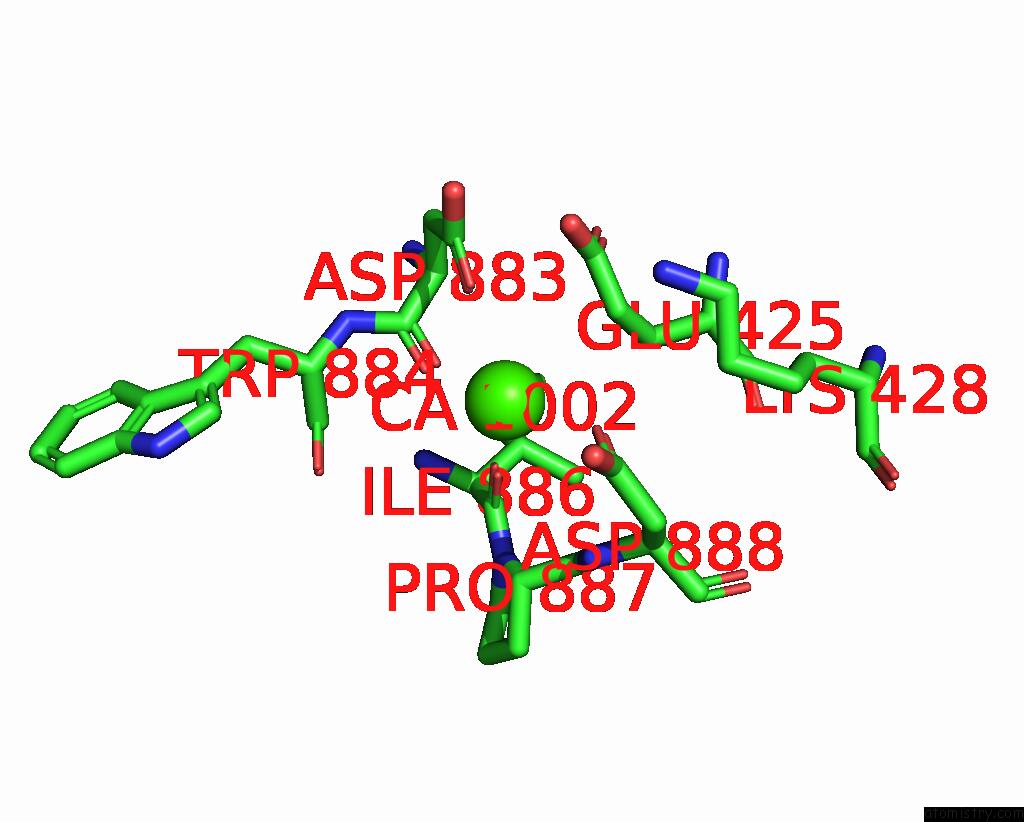
Calcium binding site 4 out of 4 in 8qzc
Go back to
Calcium binding site 4 out
of 4 in the Structure of Calcium-Bound MTMEM16A(Ac)-L647V/I733V Chloride Channel at 3.29 A Resolution
Mono view
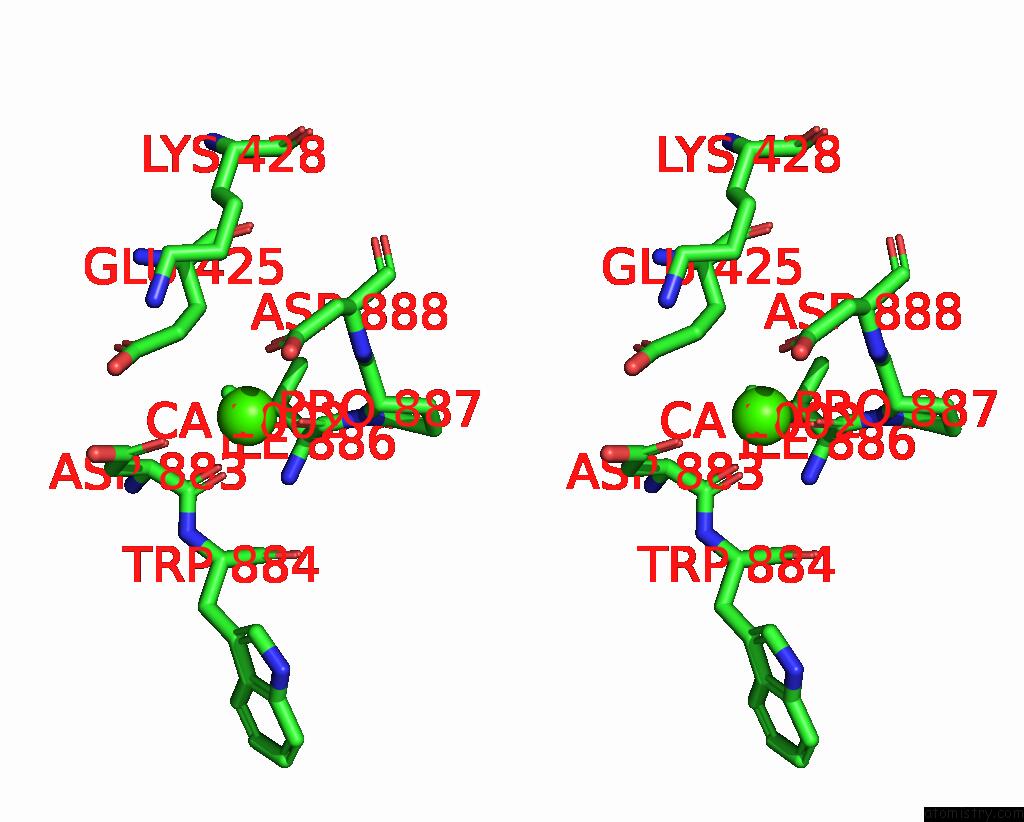
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of Structure of Calcium-Bound MTMEM16A(Ac)-L647V/I733V Chloride Channel at 3.29 A Resolution within 5.0Å range:
|
Reference:
A.K.Lam,
R.Dutzler.
Mechanistic Basis of Ligand Efficacy in the Calcium-Activated Chloride Channel TMEM16A. Embo J. 15030 2023.
ISSN: ESSN 1460-2075
PubMed: 37984335
DOI: 10.15252/EMBJ.2023115030
Page generated: Fri Jul 19 11:20:49 2024
ISSN: ESSN 1460-2075
PubMed: 37984335
DOI: 10.15252/EMBJ.2023115030
Last articles
Zn in 9JYWZn in 9IR4
Zn in 9IR3
Zn in 9GMX
Zn in 9GMW
Zn in 9JEJ
Zn in 9ERF
Zn in 9ERE
Zn in 9EGV
Zn in 9EGW