Calcium in PDB 8tqb: MGLUR3 in the Presence of the Agonist LY379268 and Pam VU6023326
Calcium Binding Sites:
The binding sites of Calcium atom in the MGLUR3 in the Presence of the Agonist LY379268 and Pam VU6023326
(pdb code 8tqb). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 4 binding sites of Calcium where determined in the MGLUR3 in the Presence of the Agonist LY379268 and Pam VU6023326, PDB code: 8tqb:
Jump to Calcium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Calcium where determined in the MGLUR3 in the Presence of the Agonist LY379268 and Pam VU6023326, PDB code: 8tqb:
Jump to Calcium binding site number: 1; 2; 3; 4;
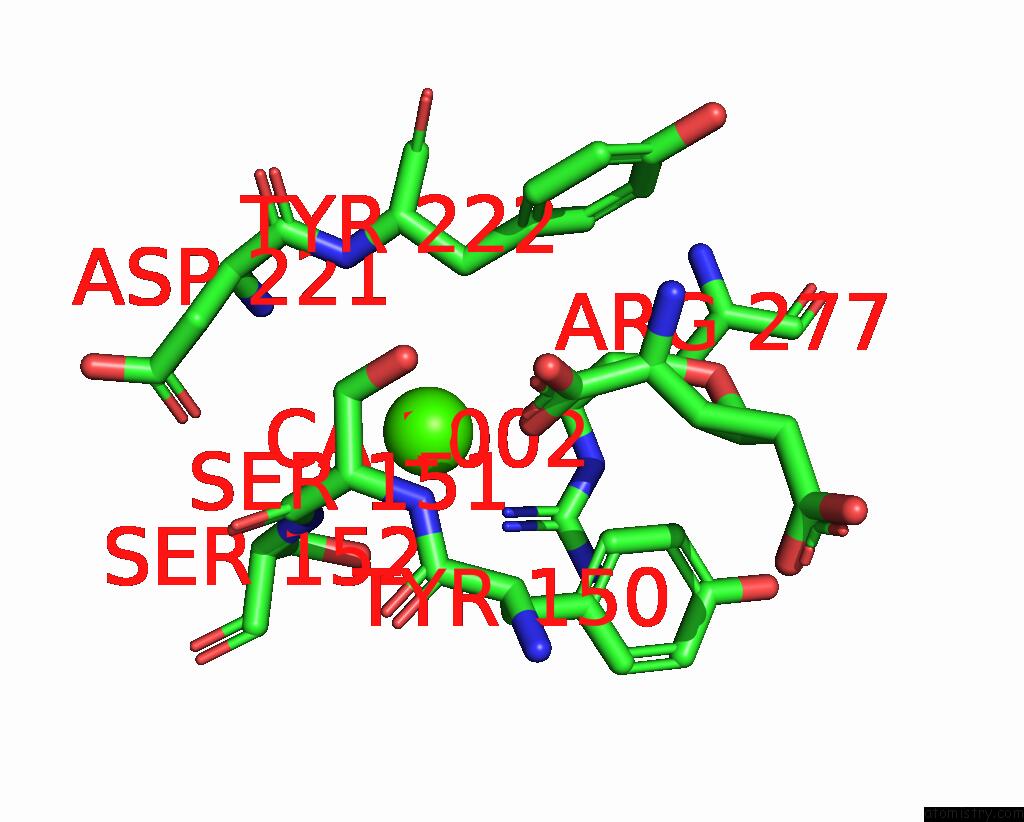
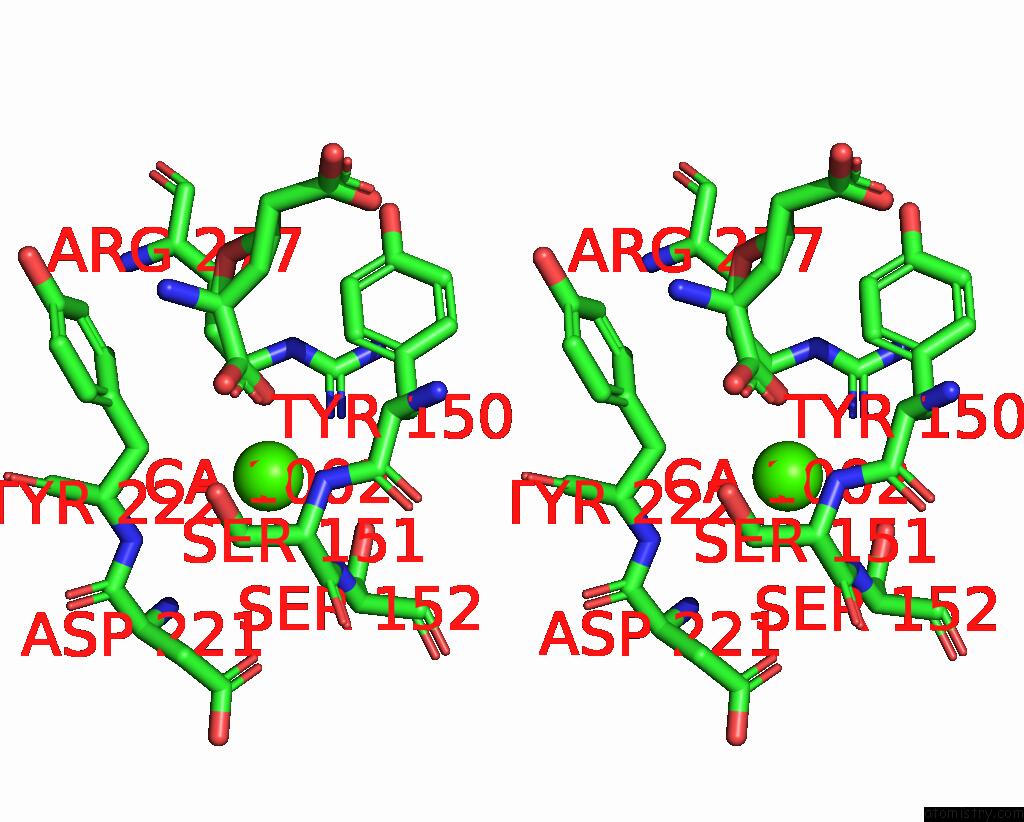
Calcium binding site 1 out of 4 in 8tqb
Go back to
Calcium binding site 1 out
of 4 in the MGLUR3 in the Presence of the Agonist LY379268 and Pam VU6023326
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of MGLUR3 in the Presence of the Agonist LY379268 and Pam VU6023326 within 5.0Å range:
|
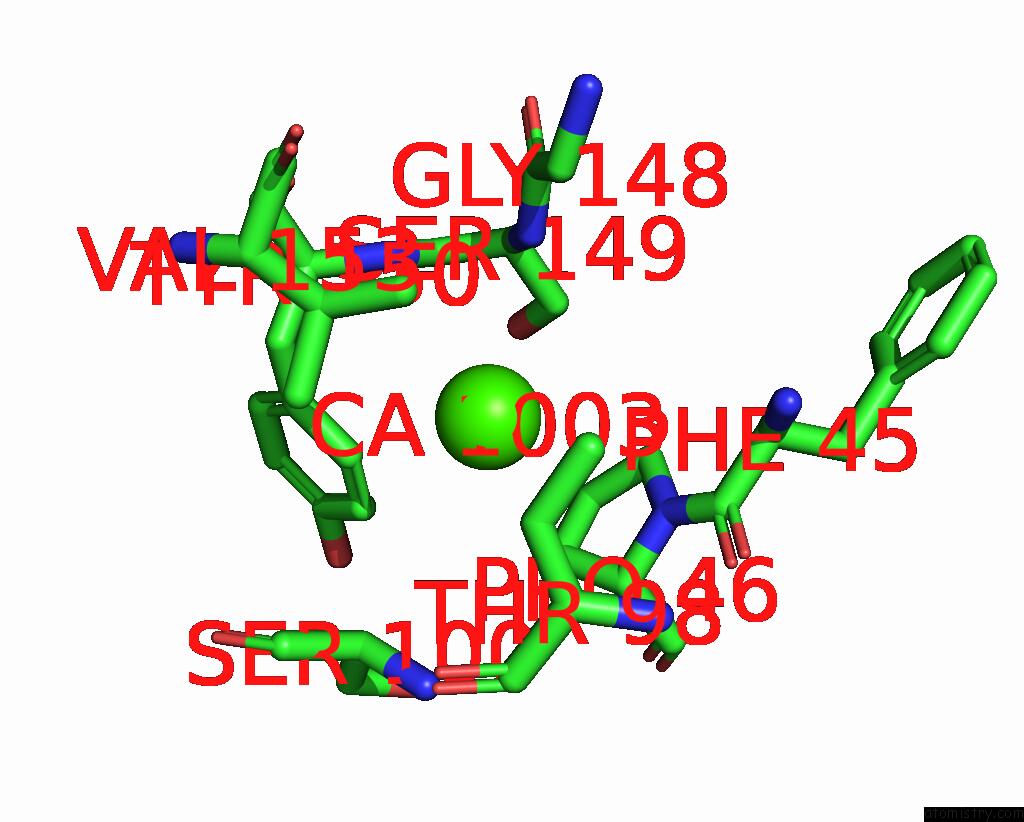
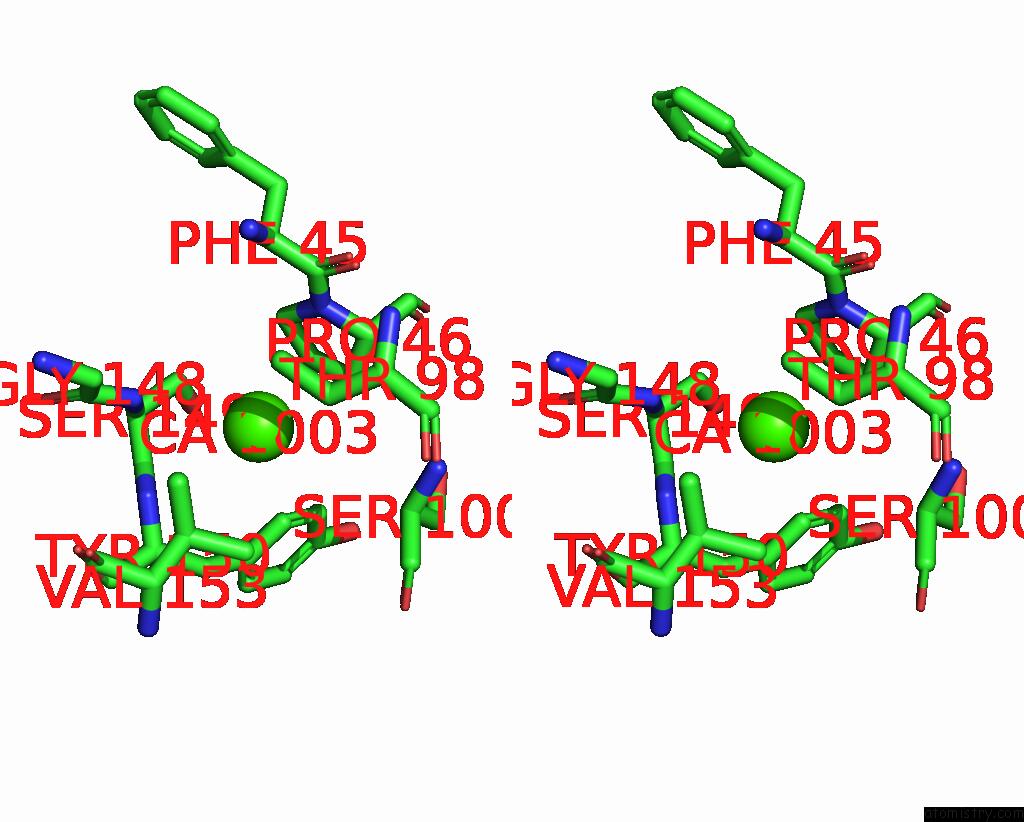
Calcium binding site 2 out of 4 in 8tqb
Go back to
Calcium binding site 2 out
of 4 in the MGLUR3 in the Presence of the Agonist LY379268 and Pam VU6023326
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of MGLUR3 in the Presence of the Agonist LY379268 and Pam VU6023326 within 5.0Å range:
|
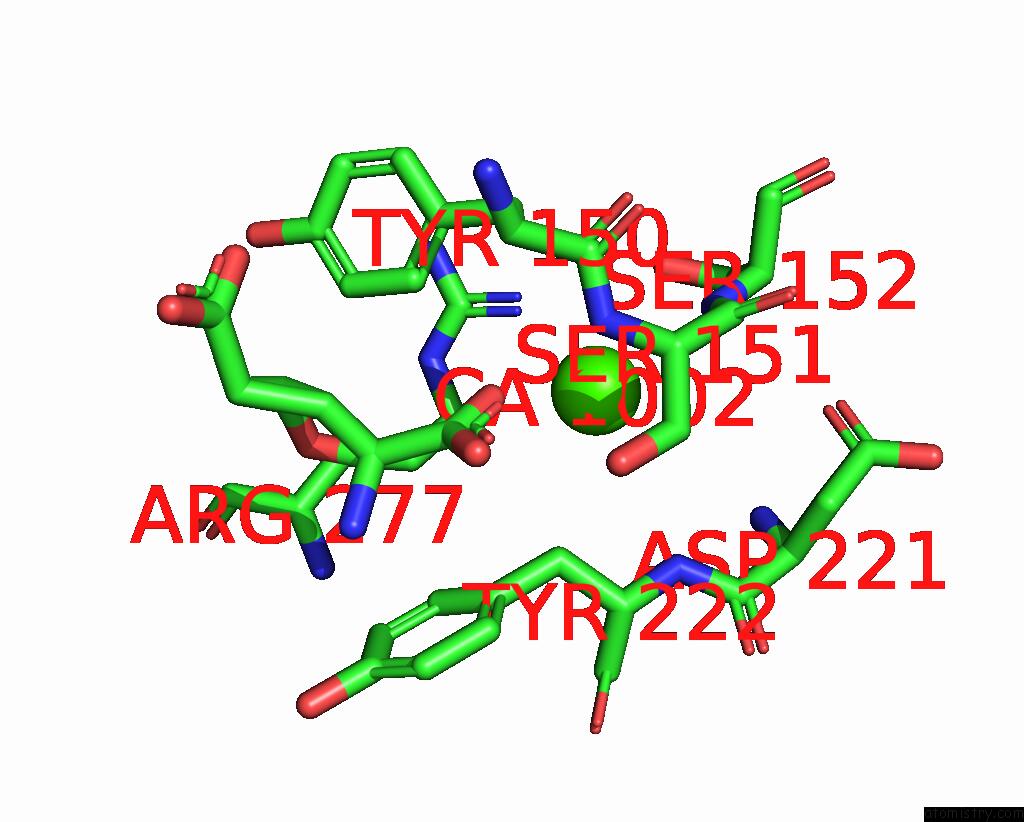
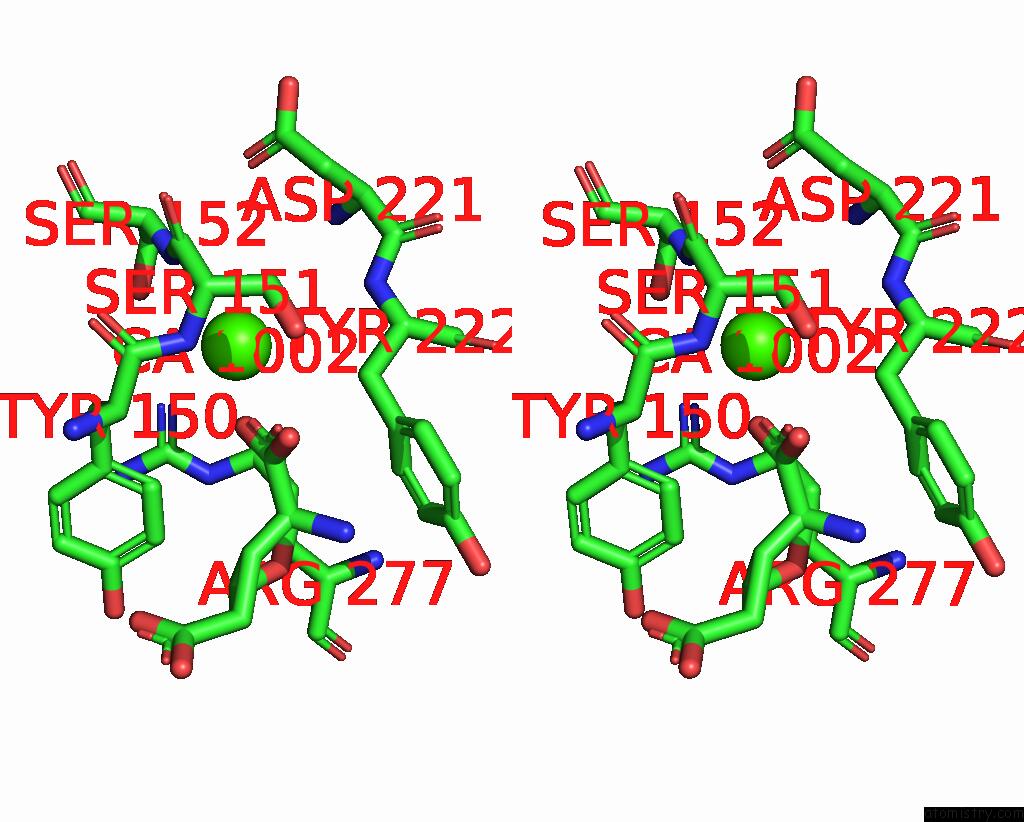
Calcium binding site 3 out of 4 in 8tqb
Go back to
Calcium binding site 3 out
of 4 in the MGLUR3 in the Presence of the Agonist LY379268 and Pam VU6023326
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of MGLUR3 in the Presence of the Agonist LY379268 and Pam VU6023326 within 5.0Å range:
|
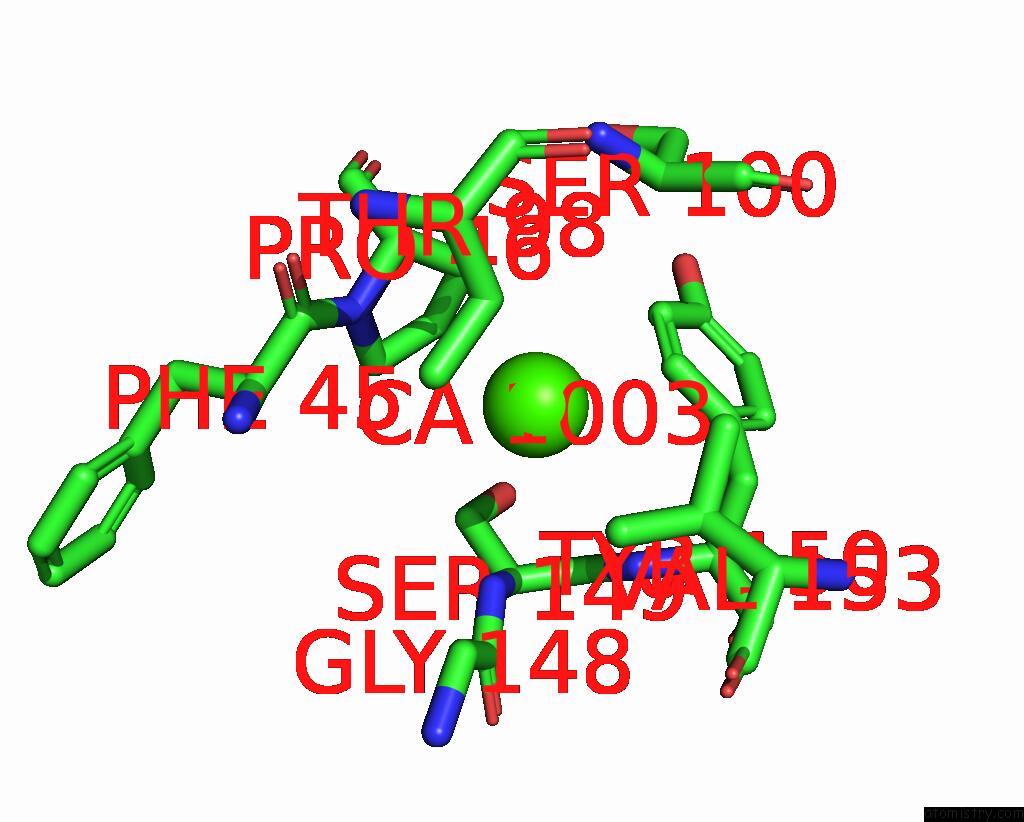
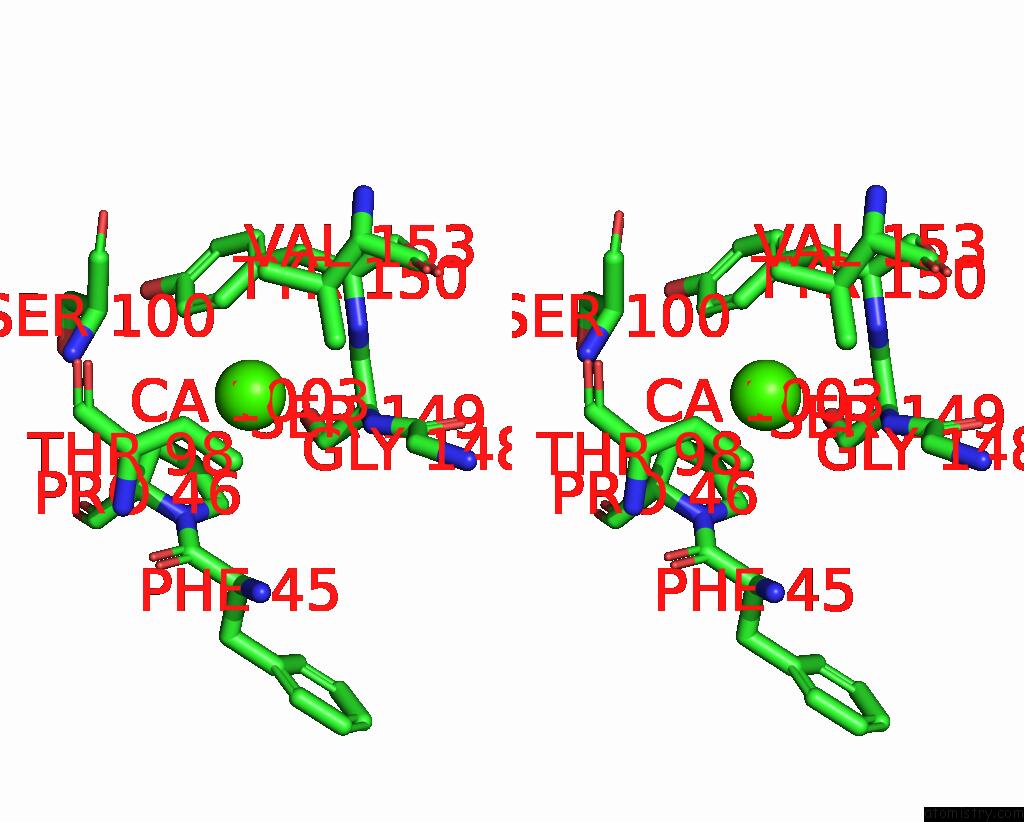
Calcium binding site 4 out of 4 in 8tqb
Go back to
Calcium binding site 4 out
of 4 in the MGLUR3 in the Presence of the Agonist LY379268 and Pam VU6023326
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of MGLUR3 in the Presence of the Agonist LY379268 and Pam VU6023326 within 5.0Å range:
|
Reference:
A.Strauss,
A.J.Gonzalez-Hernandez,
J.Lee,
N.Abreu,
P.Selvakumar,
L.Salas-Estrada,
M.Kristt,
D.C.Marx,
K.Gilliland,
B.J.Melancon,
M.Filizola,
J.Meyerson,
J.Levitz.
Structural Basis of Allosteric Modulation of Metabotropic Glutamate Receptor Activation and Desensitization. Biorxiv 2023.
ISSN: ISSN 2692-8205
PubMed: 37645747
DOI: 10.1101/2023.08.13.552748
Page generated: Sat Sep 28 09:09:05 2024
ISSN: ISSN 2692-8205
PubMed: 37645747
DOI: 10.1101/2023.08.13.552748
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1