Calcium »
PDB 1ck6-1cxi »
1cxe »
Calcium in PDB 1cxe: Complex of Cgtase with Maltotetraose at Room Temperature and pH 9.1 Based on Diffraction Data of A Crystal Soaked with Alpha-Cyclodextrin
Enzymatic activity of Complex of Cgtase with Maltotetraose at Room Temperature and pH 9.1 Based on Diffraction Data of A Crystal Soaked with Alpha-Cyclodextrin
All present enzymatic activity of Complex of Cgtase with Maltotetraose at Room Temperature and pH 9.1 Based on Diffraction Data of A Crystal Soaked with Alpha-Cyclodextrin:
2.4.1.19;
2.4.1.19;
Protein crystallography data
The structure of Complex of Cgtase with Maltotetraose at Room Temperature and pH 9.1 Based on Diffraction Data of A Crystal Soaked with Alpha-Cyclodextrin, PDB code: 1cxe
was solved by
R.M.A.Knegtel,
B.V.Strokopytov,
B.W.Dijkstra,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 8.00 / 2.10 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 121.388, 111.604, 66.386, 90.00, 90.00, 90.00 |
| R / Rfree (%) | n/a / n/a |
Calcium Binding Sites:
The binding sites of Calcium atom in the Complex of Cgtase with Maltotetraose at Room Temperature and pH 9.1 Based on Diffraction Data of A Crystal Soaked with Alpha-Cyclodextrin
(pdb code 1cxe). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Complex of Cgtase with Maltotetraose at Room Temperature and pH 9.1 Based on Diffraction Data of A Crystal Soaked with Alpha-Cyclodextrin, PDB code: 1cxe:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Complex of Cgtase with Maltotetraose at Room Temperature and pH 9.1 Based on Diffraction Data of A Crystal Soaked with Alpha-Cyclodextrin, PDB code: 1cxe:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 1cxe
Go back to
Calcium binding site 1 out
of 2 in the Complex of Cgtase with Maltotetraose at Room Temperature and pH 9.1 Based on Diffraction Data of A Crystal Soaked with Alpha-Cyclodextrin
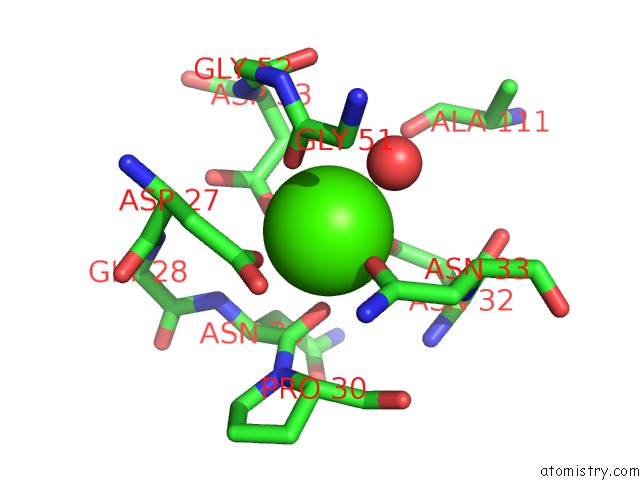
Mono view
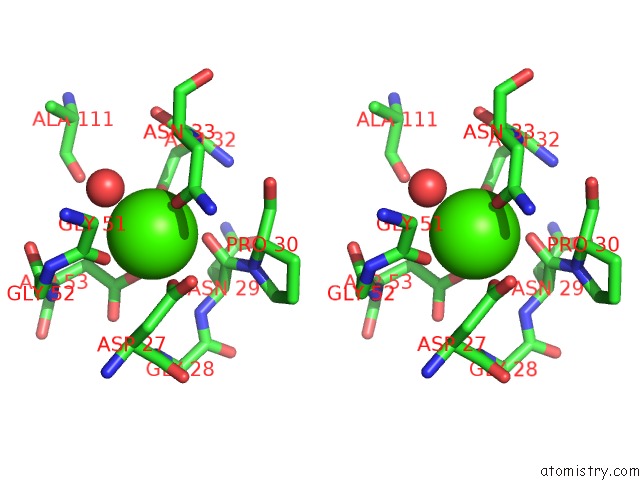
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Complex of Cgtase with Maltotetraose at Room Temperature and pH 9.1 Based on Diffraction Data of A Crystal Soaked with Alpha-Cyclodextrin within 5.0Å range:
|
Calcium binding site 2 out of 2 in 1cxe
Go back to
Calcium binding site 2 out
of 2 in the Complex of Cgtase with Maltotetraose at Room Temperature and pH 9.1 Based on Diffraction Data of A Crystal Soaked with Alpha-Cyclodextrin
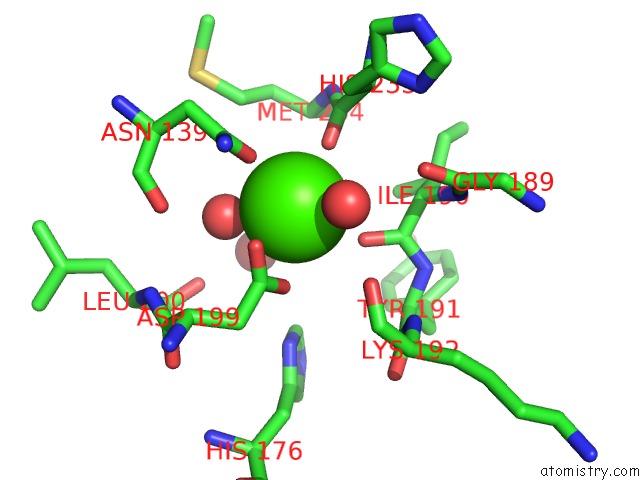
Mono view
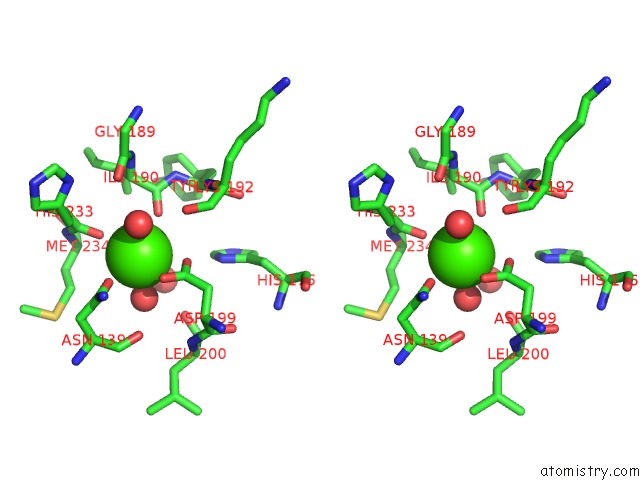
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Complex of Cgtase with Maltotetraose at Room Temperature and pH 9.1 Based on Diffraction Data of A Crystal Soaked with Alpha-Cyclodextrin within 5.0Å range:
|
Reference:
R.M.Knegtel,
B.Strokopytov,
D.Penninga,
O.G.Faber,
H.J.Rozeboom,
K.H.Kalk,
L.Dijkhuizen,
B.W.Dijkstra.
Crystallographic Studies of the Interaction of Cyclodextrin Glycosyltransferase From Bacillus Circulans Strain 251 with Natural Substrates and Products. J.Biol.Chem. V. 270 29256 1995.
ISSN: ISSN 0021-9258
PubMed: 7493956
DOI: 10.1074/JBC.270.49.29256
Page generated: Mon Jul 7 14:12:40 2025
ISSN: ISSN 0021-9258
PubMed: 7493956
DOI: 10.1074/JBC.270.49.29256
Last articles
Cl in 5KCOCl in 5KC1
Cl in 5KCN
Cl in 5KBH
Cl in 5KB6
Cl in 5KBR
Cl in 5KAI
Cl in 5KB5
Cl in 5KB3
Cl in 5KAF