Calcium »
PDB 1edm-1evu »
1eub »
Calcium in PDB 1eub: Solution Structure of the Catalytic Domain of Human Collagenase-3 (Mmp-13) Complexed to A Potent Non-Peptidic Sulfonamide Inhibitor
Other elements in 1eub:
The structure of Solution Structure of the Catalytic Domain of Human Collagenase-3 (Mmp-13) Complexed to A Potent Non-Peptidic Sulfonamide Inhibitor also contains other interesting chemical elements:
| Zinc | (Zn) | 40 atoms |
Calcium Binding Sites:
The binding sites of Calcium atom in the Solution Structure of the Catalytic Domain of Human Collagenase-3 (Mmp-13) Complexed to A Potent Non-Peptidic Sulfonamide Inhibitor
(pdb code 1eub). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Solution Structure of the Catalytic Domain of Human Collagenase-3 (Mmp-13) Complexed to A Potent Non-Peptidic Sulfonamide Inhibitor, PDB code: 1eub:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Solution Structure of the Catalytic Domain of Human Collagenase-3 (Mmp-13) Complexed to A Potent Non-Peptidic Sulfonamide Inhibitor, PDB code: 1eub:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 1eub
Go back to
Calcium binding site 1 out
of 2 in the Solution Structure of the Catalytic Domain of Human Collagenase-3 (Mmp-13) Complexed to A Potent Non-Peptidic Sulfonamide Inhibitor

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Solution Structure of the Catalytic Domain of Human Collagenase-3 (Mmp-13) Complexed to A Potent Non-Peptidic Sulfonamide Inhibitor within 5.0Å range:
|
Calcium binding site 2 out of 2 in 1eub
Go back to
Calcium binding site 2 out
of 2 in the Solution Structure of the Catalytic Domain of Human Collagenase-3 (Mmp-13) Complexed to A Potent Non-Peptidic Sulfonamide Inhibitor
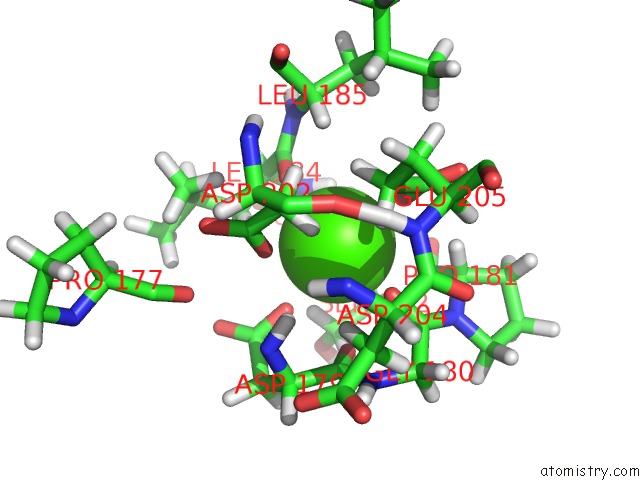
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Solution Structure of the Catalytic Domain of Human Collagenase-3 (Mmp-13) Complexed to A Potent Non-Peptidic Sulfonamide Inhibitor within 5.0Å range:
|
Reference:
X.Zhang,
N.C.Gonnella,
J.Koehn,
N.Pathak,
V.Ganu,
R.Melton,
D.Parker,
S.I.Hu,
K.Y.Nam.
Solution Structure of the Catalytic Domain of Human Collagenase-3 (Mmp-13) Complexed to A Potent Non-Peptidic Sulfonamide Inhibitor: Binding Comparison with Stromelysin-1 and Collagenase-1. J.Mol.Biol. V. 301 513 2000.
ISSN: ISSN 0022-2836
PubMed: 10926524
DOI: 10.1006/JMBI.2000.3988
Page generated: Mon Jul 7 14:43:00 2025
ISSN: ISSN 0022-2836
PubMed: 10926524
DOI: 10.1006/JMBI.2000.3988
Last articles
Ca in 7G7NCa in 7G7K
Ca in 7G7M
Ca in 7G7J
Ca in 7G7I
Ca in 7G7G
Ca in 7G7H
Ca in 7G7F
Ca in 7G7E
Ca in 7G7C