Calcium »
PDB 1hov-1i6t »
1i4a »
Calcium in PDB 1i4a: Crystal Structure of Phosphorylation-Mimicking Mutant T6D of Annexin IV
Protein crystallography data
The structure of Crystal Structure of Phosphorylation-Mimicking Mutant T6D of Annexin IV, PDB code: 1i4a
was solved by
M.A.Kaetzel,
Y.D.Mo,
T.R.Mealy,
B.Campos,
W.Bergsma-Schutter,
A.Brisson,
J.R.Dedman,
B.A.Seaton,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 100.00 / 2.00 |
| Space group | H 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 119.060, 119.060, 82.160, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 20.4 / 23 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of Phosphorylation-Mimicking Mutant T6D of Annexin IV
(pdb code 1i4a). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Crystal Structure of Phosphorylation-Mimicking Mutant T6D of Annexin IV, PDB code: 1i4a:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Crystal Structure of Phosphorylation-Mimicking Mutant T6D of Annexin IV, PDB code: 1i4a:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 1i4a
Go back to
Calcium binding site 1 out
of 2 in the Crystal Structure of Phosphorylation-Mimicking Mutant T6D of Annexin IV
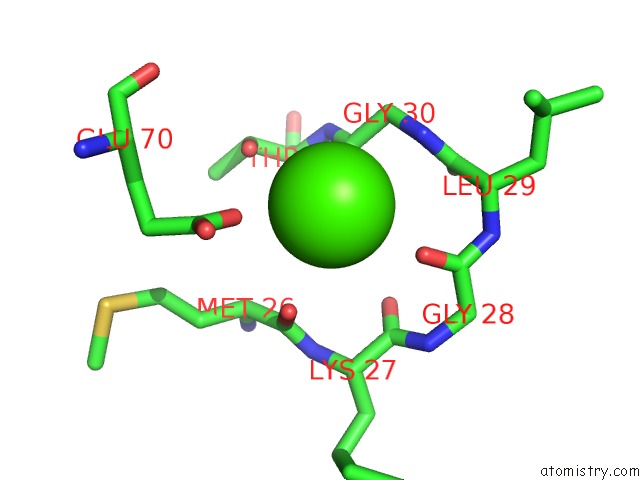
Mono view
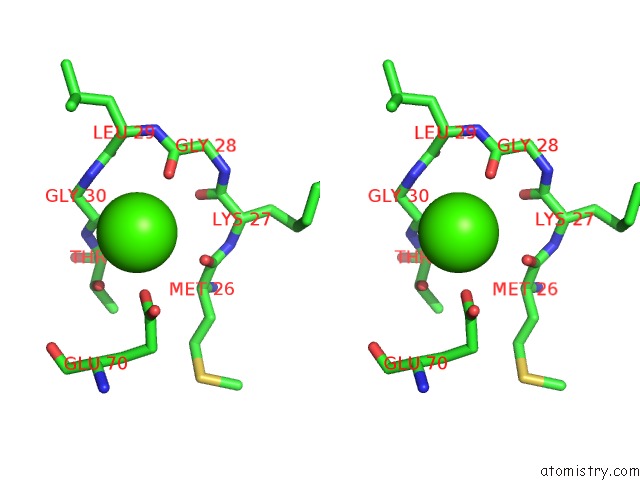
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of Phosphorylation-Mimicking Mutant T6D of Annexin IV within 5.0Å range:
|
Calcium binding site 2 out of 2 in 1i4a
Go back to
Calcium binding site 2 out
of 2 in the Crystal Structure of Phosphorylation-Mimicking Mutant T6D of Annexin IV
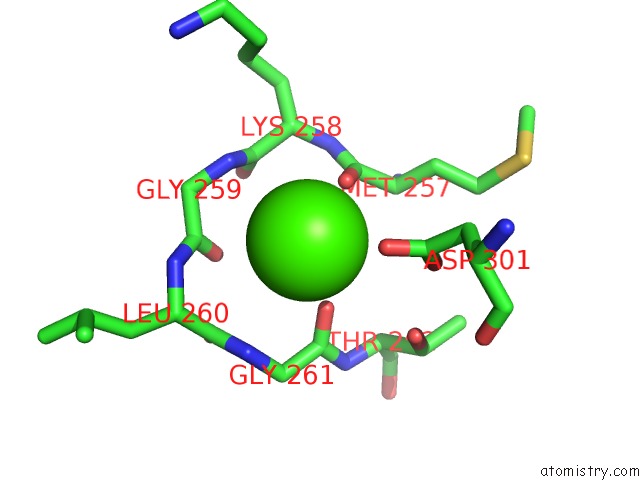
Mono view
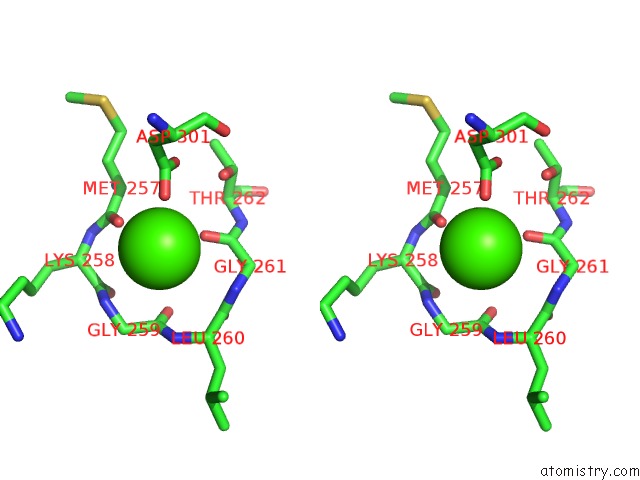
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Crystal Structure of Phosphorylation-Mimicking Mutant T6D of Annexin IV within 5.0Å range:
|
Reference:
M.A.Kaetzel,
Y.D.Mo,
T.R.Mealy,
B.Campos,
W.Bergsma-Schutter,
A.Brisson,
J.R.Dedman,
B.A.Seaton.
Phosphorylation Mutants Elucidate the Mechanism of Annexin IV-Mediated Membrane Aggregation. Biochemistry V. 40 4192 2001.
ISSN: ISSN 0006-2960
PubMed: 11300800
DOI: 10.1021/BI002507S
Page generated: Mon Jul 7 15:46:27 2025
ISSN: ISSN 0006-2960
PubMed: 11300800
DOI: 10.1021/BI002507S
Last articles
Cl in 5JZSCl in 5K0E
Cl in 5JY3
Cl in 5JZN
Cl in 5JZB
Cl in 5JZL
Cl in 5JZ9
Cl in 5JZK
Cl in 5JY1
Cl in 5JYL