Calcium »
PDB 1nya-1o3c »
1o0q »
Calcium in PDB 1o0q: Crystal Structure of A Cold Adapted Alkaline Protease From Pseudomonas Tac II 18, Co-Crystallized with 1 Mm Edta
Enzymatic activity of Crystal Structure of A Cold Adapted Alkaline Protease From Pseudomonas Tac II 18, Co-Crystallized with 1 Mm Edta
All present enzymatic activity of Crystal Structure of A Cold Adapted Alkaline Protease From Pseudomonas Tac II 18, Co-Crystallized with 1 Mm Edta:
3.4.24.40;
3.4.24.40;
Protein crystallography data
The structure of Crystal Structure of A Cold Adapted Alkaline Protease From Pseudomonas Tac II 18, Co-Crystallized with 1 Mm Edta, PDB code: 1o0q
was solved by
S.Ravaud,
P.Gouet,
R.Haser,
N.Aghajari,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.20 |
| Space group | H 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 179.900, 179.900, 37.300, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 18.9 / 23.7 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of A Cold Adapted Alkaline Protease From Pseudomonas Tac II 18, Co-Crystallized with 1 Mm Edta
(pdb code 1o0q). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 6 binding sites of Calcium where determined in the Crystal Structure of A Cold Adapted Alkaline Protease From Pseudomonas Tac II 18, Co-Crystallized with 1 Mm Edta, PDB code: 1o0q:
Jump to Calcium binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Calcium where determined in the Crystal Structure of A Cold Adapted Alkaline Protease From Pseudomonas Tac II 18, Co-Crystallized with 1 Mm Edta, PDB code: 1o0q:
Jump to Calcium binding site number: 1; 2; 3; 4; 5; 6;
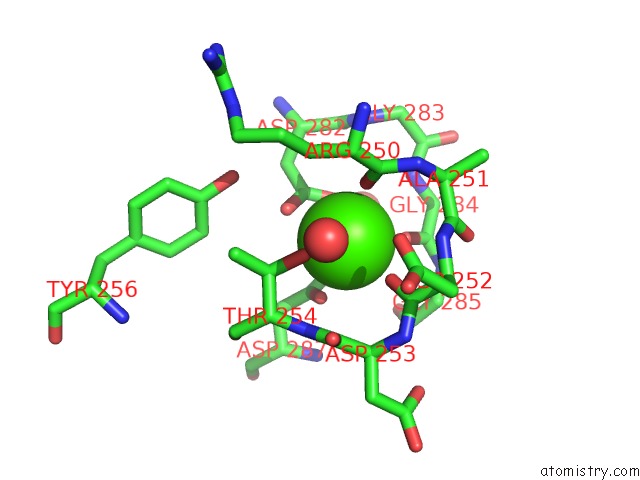
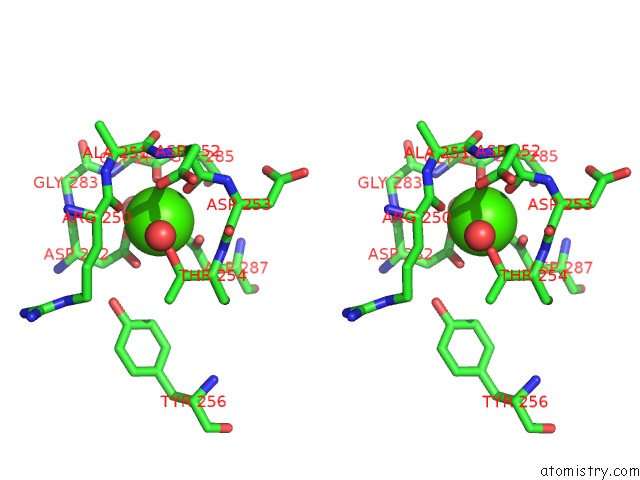
Calcium binding site 1 out of 6 in 1o0q
Go back to
Calcium binding site 1 out
of 6 in the Crystal Structure of A Cold Adapted Alkaline Protease From Pseudomonas Tac II 18, Co-Crystallized with 1 Mm Edta
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of A Cold Adapted Alkaline Protease From Pseudomonas Tac II 18, Co-Crystallized with 1 Mm Edta within 5.0Å range:
|
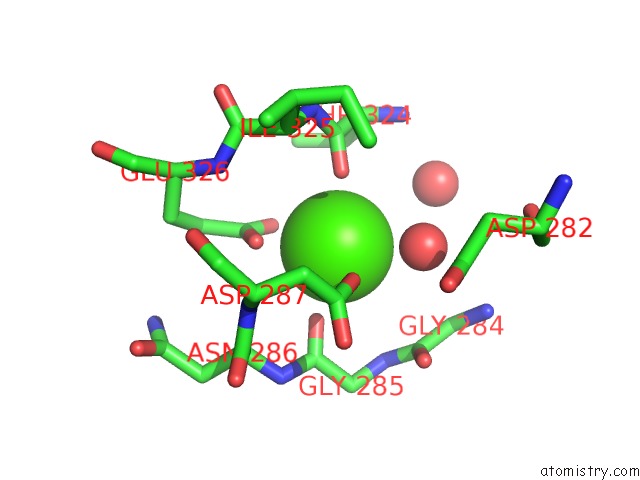
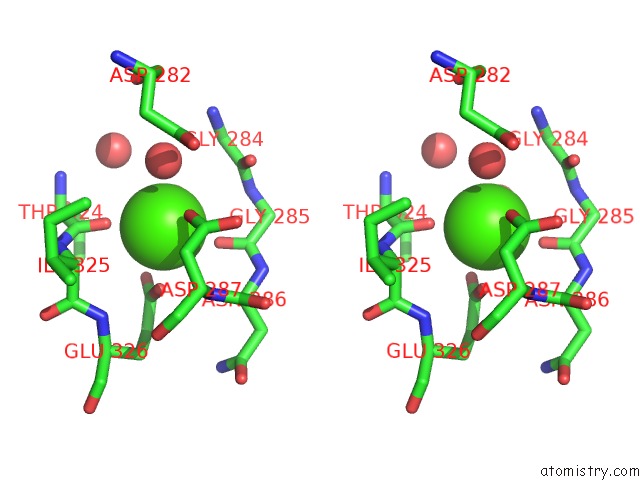
Calcium binding site 2 out of 6 in 1o0q
Go back to
Calcium binding site 2 out
of 6 in the Crystal Structure of A Cold Adapted Alkaline Protease From Pseudomonas Tac II 18, Co-Crystallized with 1 Mm Edta
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Crystal Structure of A Cold Adapted Alkaline Protease From Pseudomonas Tac II 18, Co-Crystallized with 1 Mm Edta within 5.0Å range:
|
Calcium binding site 3 out of 6 in 1o0q
Go back to
Calcium binding site 3 out
of 6 in the Crystal Structure of A Cold Adapted Alkaline Protease From Pseudomonas Tac II 18, Co-Crystallized with 1 Mm Edta
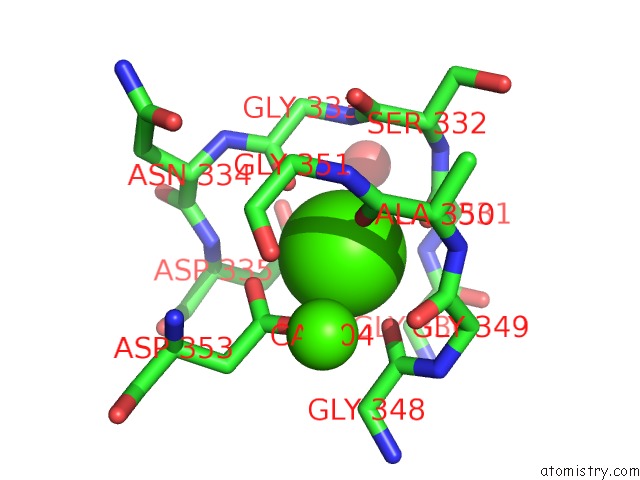
Mono view
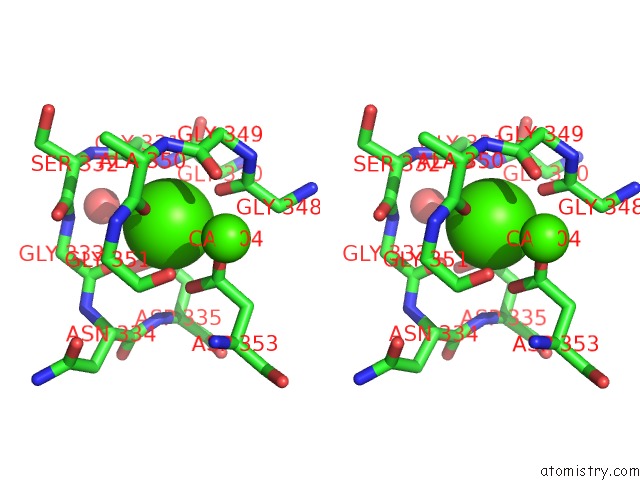
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Crystal Structure of A Cold Adapted Alkaline Protease From Pseudomonas Tac II 18, Co-Crystallized with 1 Mm Edta within 5.0Å range:
|
Calcium binding site 4 out of 6 in 1o0q
Go back to
Calcium binding site 4 out
of 6 in the Crystal Structure of A Cold Adapted Alkaline Protease From Pseudomonas Tac II 18, Co-Crystallized with 1 Mm Edta
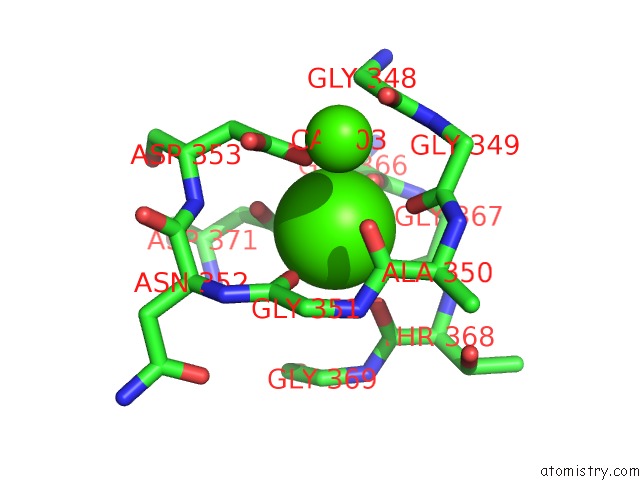
Mono view
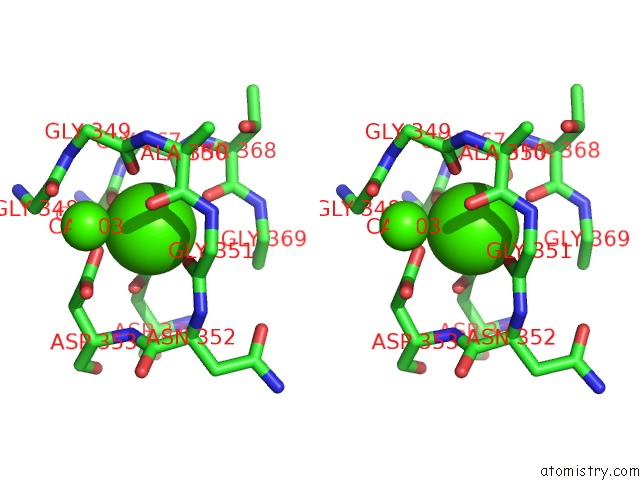
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of Crystal Structure of A Cold Adapted Alkaline Protease From Pseudomonas Tac II 18, Co-Crystallized with 1 Mm Edta within 5.0Å range:
|
Calcium binding site 5 out of 6 in 1o0q
Go back to
Calcium binding site 5 out
of 6 in the Crystal Structure of A Cold Adapted Alkaline Protease From Pseudomonas Tac II 18, Co-Crystallized with 1 Mm Edta
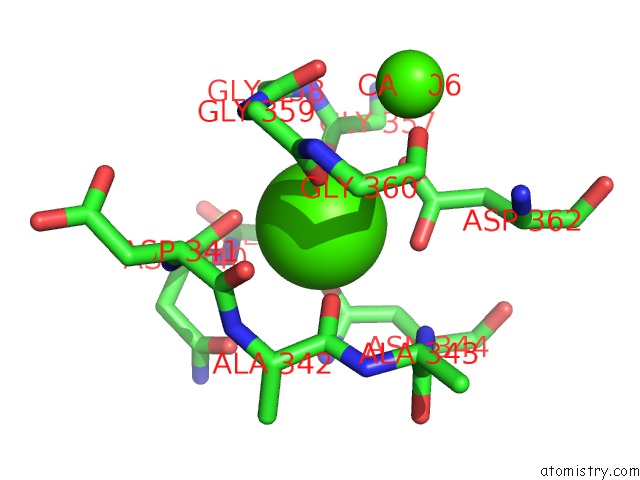
Mono view
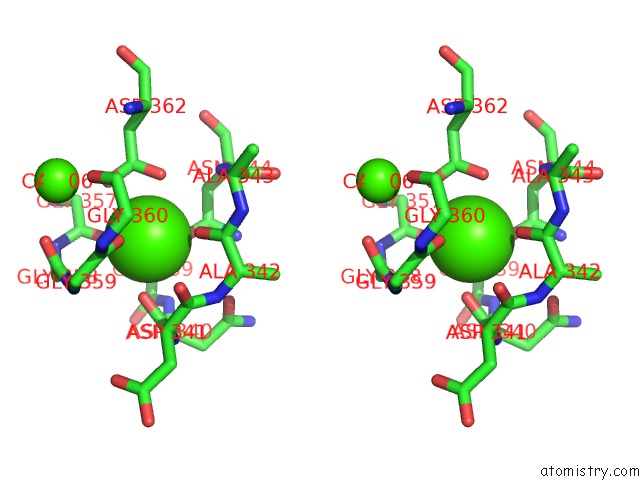
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 5 of Crystal Structure of A Cold Adapted Alkaline Protease From Pseudomonas Tac II 18, Co-Crystallized with 1 Mm Edta within 5.0Å range:
|
Calcium binding site 6 out of 6 in 1o0q
Go back to
Calcium binding site 6 out
of 6 in the Crystal Structure of A Cold Adapted Alkaline Protease From Pseudomonas Tac II 18, Co-Crystallized with 1 Mm Edta
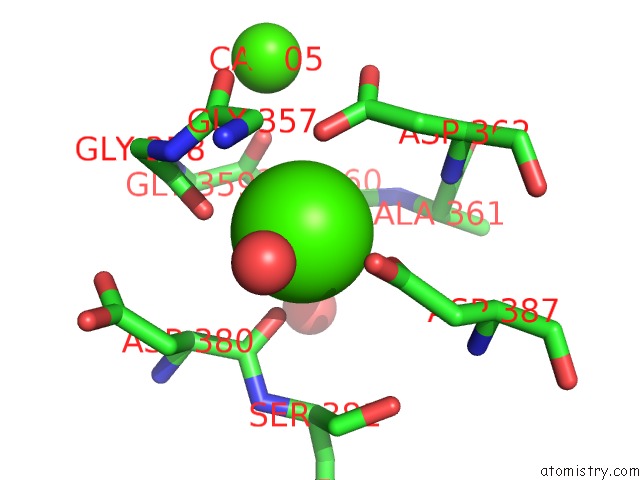
Mono view
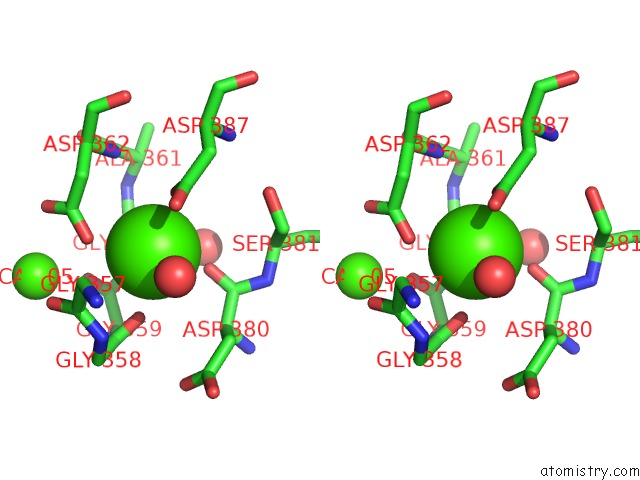
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 6 of Crystal Structure of A Cold Adapted Alkaline Protease From Pseudomonas Tac II 18, Co-Crystallized with 1 Mm Edta within 5.0Å range:
|
Reference:
S.Ravaud,
P.Gouet,
R.Haser,
N.Aghajari.
Probing the Role of Divalent Metal Ions in A Bacterial Psychrophilic Metalloprotease: Binding Studies of An Enzyme in the Crystalline State By X-Ray Crystallography. J.Bacteriol. V. 185 4195 2003.
ISSN: ISSN 0021-9193
PubMed: 12837794
DOI: 10.1128/JB.185.14.4195-4203.2003
Page generated: Mon Jul 7 17:44:43 2025
ISSN: ISSN 0021-9193
PubMed: 12837794
DOI: 10.1128/JB.185.14.4195-4203.2003
Last articles
Cl in 5KGMCl in 5KGR
Cl in 5KH3
Cl in 5KGT
Cl in 5KGN
Cl in 5KGG
Cl in 5KGI
Cl in 5KGL
Cl in 5KGH
Cl in 5KC9