Calcium »
PDB 1pig-1px2 »
1prs »
Calcium in PDB 1prs: uc(Nmr)-Derived Three-Dimensional Solution Structure of Protein S Complexed with Calcium
Calcium Binding Sites:
The binding sites of Calcium atom in the uc(Nmr)-Derived Three-Dimensional Solution Structure of Protein S Complexed with Calcium
(pdb code 1prs). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the uc(Nmr)-Derived Three-Dimensional Solution Structure of Protein S Complexed with Calcium, PDB code: 1prs:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the uc(Nmr)-Derived Three-Dimensional Solution Structure of Protein S Complexed with Calcium, PDB code: 1prs:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 1prs
Go back to
Calcium binding site 1 out
of 2 in the uc(Nmr)-Derived Three-Dimensional Solution Structure of Protein S Complexed with Calcium
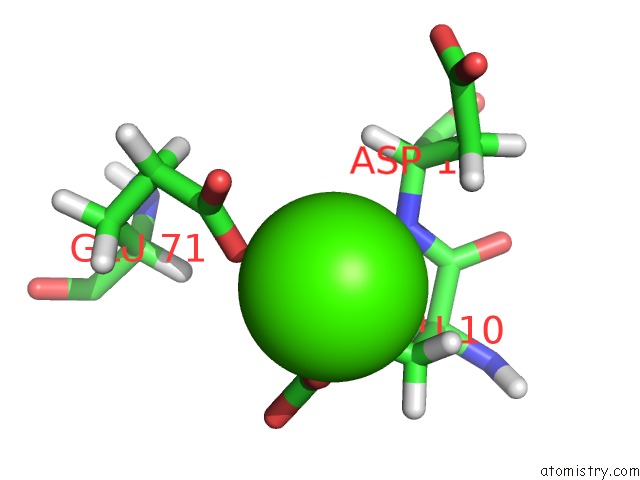
Mono view
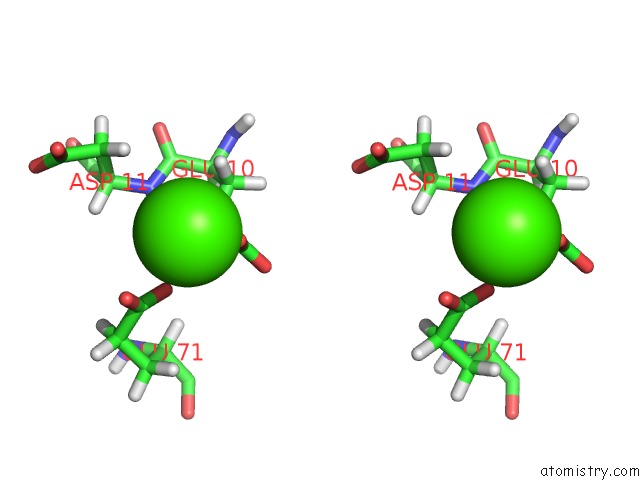
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of uc(Nmr)-Derived Three-Dimensional Solution Structure of Protein S Complexed with Calcium within 5.0Å range:
|
Calcium binding site 2 out of 2 in 1prs
Go back to
Calcium binding site 2 out
of 2 in the uc(Nmr)-Derived Three-Dimensional Solution Structure of Protein S Complexed with Calcium
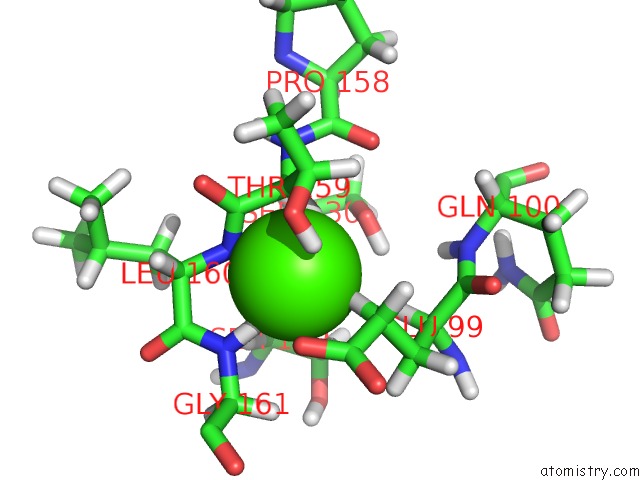
Mono view
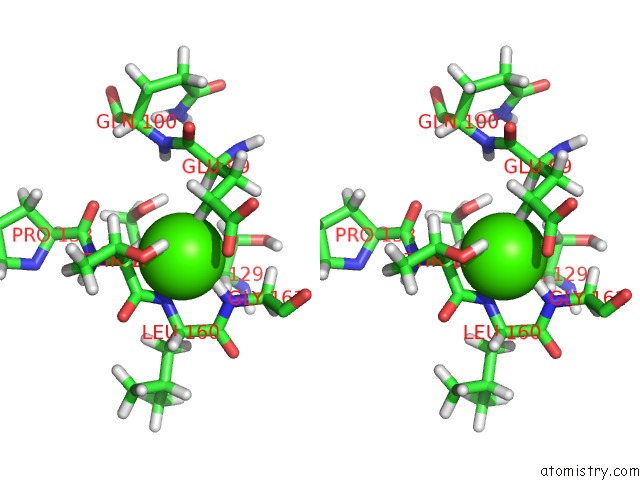
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of uc(Nmr)-Derived Three-Dimensional Solution Structure of Protein S Complexed with Calcium within 5.0Å range:
|
Reference:
S.Bagby,
T.S.Harvey,
S.G.Eagle,
S.Inouye,
M.Ikura.
uc(Nmr)-Derived Three-Dimensional Solution Structure of Protein S Complexed with Calcium. Structure V. 2 107 1994.
ISSN: ISSN 0969-2126
PubMed: 8081742
DOI: 10.1016/S0969-2126(00)00013-7
Page generated: Thu Jul 11 13:52:10 2024
ISSN: ISSN 0969-2126
PubMed: 8081742
DOI: 10.1016/S0969-2126(00)00013-7
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1