Calcium »
PDB 1qd6-1qlk »
1qd6 »
Calcium in PDB 1qd6: Outer Membrane Phospholipase A From Escherichia Coli
Enzymatic activity of Outer Membrane Phospholipase A From Escherichia Coli
All present enzymatic activity of Outer Membrane Phospholipase A From Escherichia Coli:
3.1.1.32;
3.1.1.32;
Protein crystallography data
The structure of Outer Membrane Phospholipase A From Escherichia Coli, PDB code: 1qd6
was solved by
H.J.Snijder,
I.Ubarretxena-Belandia,
M.Blaauw,
K.H.Kalk,
H.M.Verheij,
M.R.Egmond,
N.Dekker,
B.W.Dijkstra,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.30 / 2.10 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 80.067, 84.038, 95.245, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 22.6 / 28.3 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Outer Membrane Phospholipase A From Escherichia Coli
(pdb code 1qd6). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Outer Membrane Phospholipase A From Escherichia Coli, PDB code: 1qd6:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Outer Membrane Phospholipase A From Escherichia Coli, PDB code: 1qd6:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 1qd6
Go back to
Calcium binding site 1 out
of 2 in the Outer Membrane Phospholipase A From Escherichia Coli
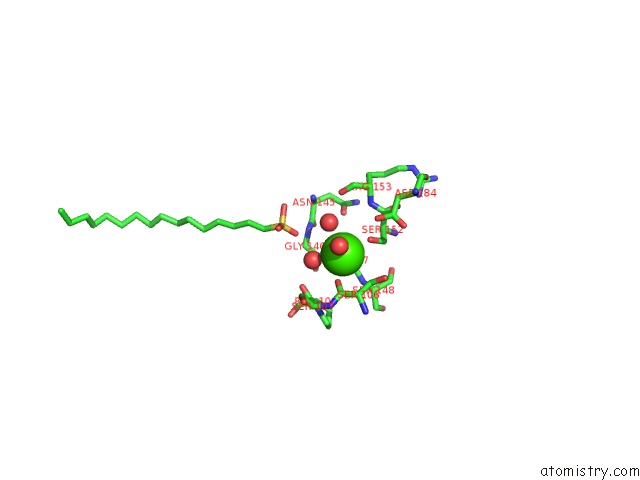
Mono view
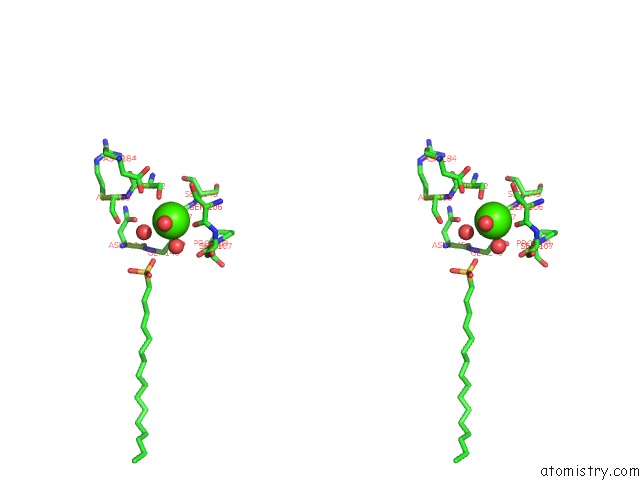
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Outer Membrane Phospholipase A From Escherichia Coli within 5.0Å range:
|
Calcium binding site 2 out of 2 in 1qd6
Go back to
Calcium binding site 2 out
of 2 in the Outer Membrane Phospholipase A From Escherichia Coli
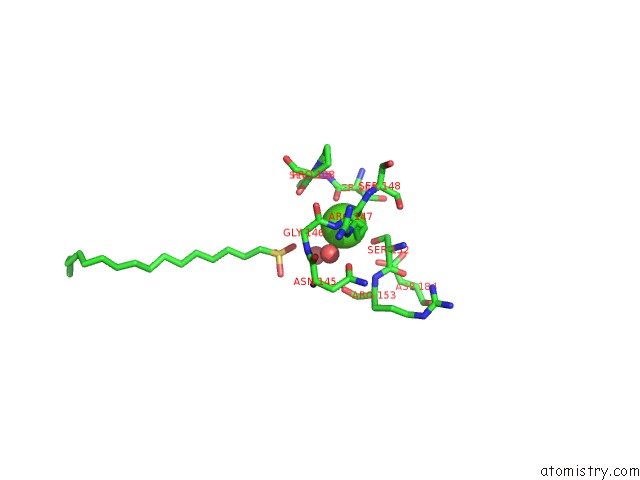
Mono view
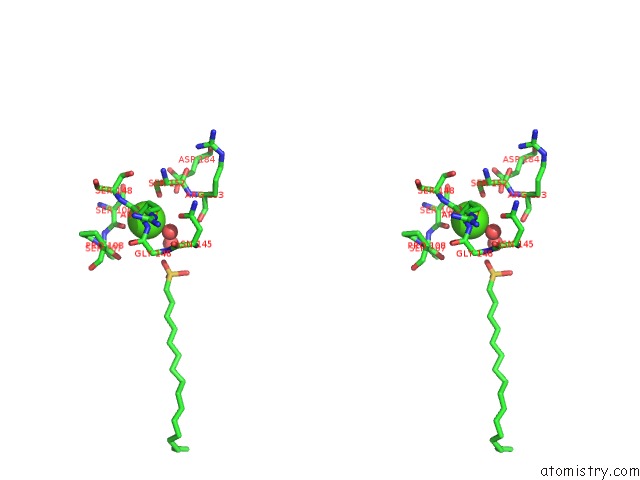
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Outer Membrane Phospholipase A From Escherichia Coli within 5.0Å range:
|
Reference:
H.J.Snijder,
I.Ubarretxena-Belandia,
M.Blaauw,
K.H.Kalk,
H.M.Verheij,
M.R.Egmond,
N.Dekker,
B.W.Dijkstra.
Structural Evidence For Dimerization-Regulated Activation of An Integral Membrane Phospholipase. Nature V. 401 717 1999.
ISSN: ISSN 0028-0836
PubMed: 10537112
DOI: 10.1038/44890
Page generated: Thu Jul 11 21:42:24 2024
ISSN: ISSN 0028-0836
PubMed: 10537112
DOI: 10.1038/44890
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1