Calcium »
PDB 1qls-1ra1 »
1ra1 »
Calcium in PDB 1ra1: Dihydrofolate Reductase Complexed with Nicotinamide Adenine Dinucleotide Phosphate (Reduced Form)
Enzymatic activity of Dihydrofolate Reductase Complexed with Nicotinamide Adenine Dinucleotide Phosphate (Reduced Form)
All present enzymatic activity of Dihydrofolate Reductase Complexed with Nicotinamide Adenine Dinucleotide Phosphate (Reduced Form):
1.5.1.3;
1.5.1.3;
Protein crystallography data
The structure of Dihydrofolate Reductase Complexed with Nicotinamide Adenine Dinucleotide Phosphate (Reduced Form), PDB code: 1ra1
was solved by
M.R.Sawaya,
J.Kraut,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 1.90 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 74.890, 59.230, 38.900, 90.00, 106.27, 90.00 |
| R / Rfree (%) | 14.7 / n/a |
Calcium Binding Sites:
The binding sites of Calcium atom in the Dihydrofolate Reductase Complexed with Nicotinamide Adenine Dinucleotide Phosphate (Reduced Form)
(pdb code 1ra1). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total only one binding site of Calcium was determined in the Dihydrofolate Reductase Complexed with Nicotinamide Adenine Dinucleotide Phosphate (Reduced Form), PDB code: 1ra1:
In total only one binding site of Calcium was determined in the Dihydrofolate Reductase Complexed with Nicotinamide Adenine Dinucleotide Phosphate (Reduced Form), PDB code: 1ra1:
Calcium binding site 1 out of 1 in 1ra1
Go back to
Calcium binding site 1 out
of 1 in the Dihydrofolate Reductase Complexed with Nicotinamide Adenine Dinucleotide Phosphate (Reduced Form)
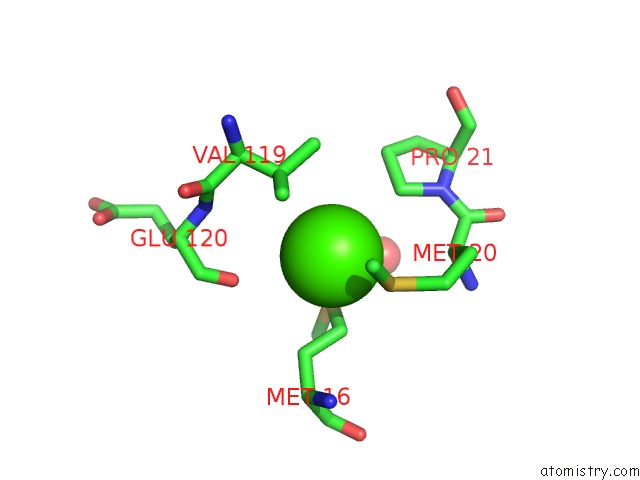
Mono view
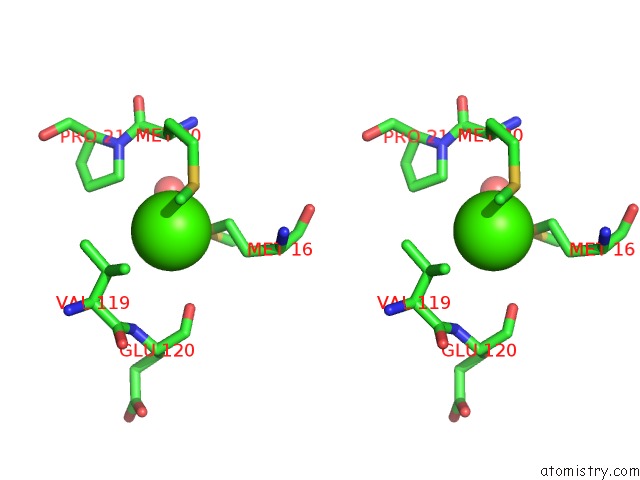
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Dihydrofolate Reductase Complexed with Nicotinamide Adenine Dinucleotide Phosphate (Reduced Form) within 5.0Å range:
|
Reference:
M.R.Sawaya,
J.Kraut.
Loop and Subdomain Movements in the Mechanism of Escherichia Coli Dihydrofolate Reductase: Crystallographic Evidence. Biochemistry V. 36 586 1997.
ISSN: ISSN 0006-2960
PubMed: 9012674
DOI: 10.1021/BI962337C
Page generated: Tue Jul 8 01:33:17 2025
ISSN: ISSN 0006-2960
PubMed: 9012674
DOI: 10.1021/BI962337C
Last articles
Cl in 5JXKCl in 5JXR
Cl in 5JXP
Cl in 5JXQ
Cl in 5JXJ
Cl in 5JX0
Cl in 5JXF
Cl in 5JX3
Cl in 5JXI
Cl in 5JXH