Calcium »
PDB 1wy9-1xjo »
1wyg »
Calcium in PDB 1wyg: Crystal Structure of A Rat Xanthine Dehydrogenase Triple Mutant (C535A, C992R and C1324S)
Enzymatic activity of Crystal Structure of A Rat Xanthine Dehydrogenase Triple Mutant (C535A, C992R and C1324S)
All present enzymatic activity of Crystal Structure of A Rat Xanthine Dehydrogenase Triple Mutant (C535A, C992R and C1324S):
1.1.1.204; 1.1.3.22;
1.1.1.204; 1.1.3.22;
Protein crystallography data
The structure of Crystal Structure of A Rat Xanthine Dehydrogenase Triple Mutant (C535A, C992R and C1324S), PDB code: 1wyg
was solved by
T.Nishino,
K.Okamoto,
Y.Kawaguchi,
H.Hori,
T.Matsumura,
B.T.Eger,
E.F.Pai,
T.Nishino,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 25.00 / 2.60 |
| Space group | I 41 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 134.253, 134.253, 523.315, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.3 / 24.8 |
Other elements in 1wyg:
The structure of Crystal Structure of A Rat Xanthine Dehydrogenase Triple Mutant (C535A, C992R and C1324S) also contains other interesting chemical elements:
| Iron | (Fe) | 4 atoms |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of A Rat Xanthine Dehydrogenase Triple Mutant (C535A, C992R and C1324S)
(pdb code 1wyg). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total only one binding site of Calcium was determined in the Crystal Structure of A Rat Xanthine Dehydrogenase Triple Mutant (C535A, C992R and C1324S), PDB code: 1wyg:
In total only one binding site of Calcium was determined in the Crystal Structure of A Rat Xanthine Dehydrogenase Triple Mutant (C535A, C992R and C1324S), PDB code: 1wyg:
Calcium binding site 1 out of 1 in 1wyg
Go back to
Calcium binding site 1 out
of 1 in the Crystal Structure of A Rat Xanthine Dehydrogenase Triple Mutant (C535A, C992R and C1324S)
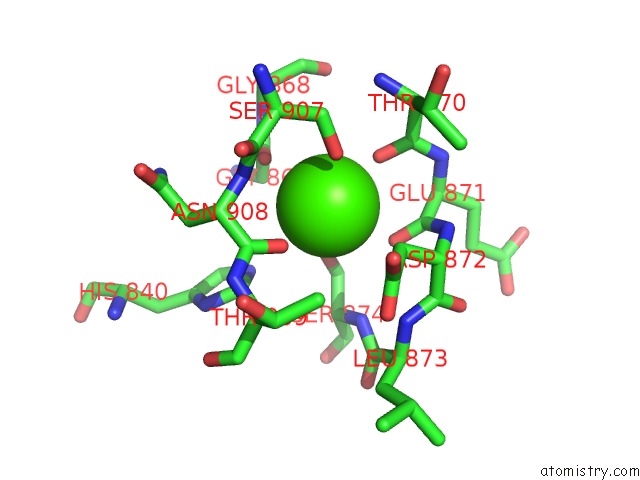
Mono view
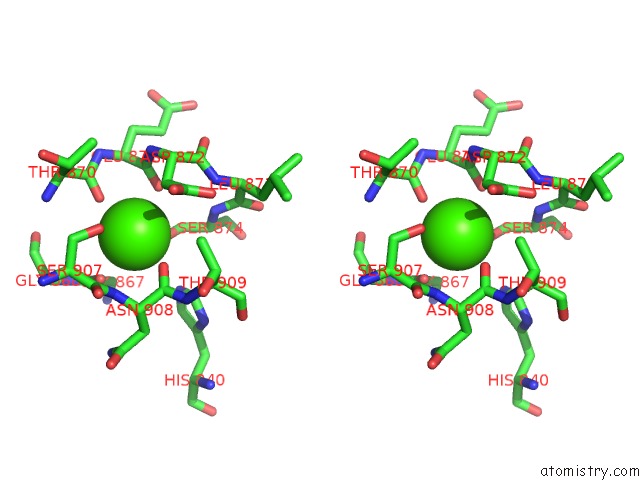
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of A Rat Xanthine Dehydrogenase Triple Mutant (C535A, C992R and C1324S) within 5.0Å range:
|
Reference:
T.Nishino,
K.Okamoto,
Y.Kawaguchi,
H.Hori,
T.Matsumura,
B.T.Eger,
E.F.Pai,
T.Nishino.
Mechanism of the Conversion of Xanthine Dehydrogenase to Xanthine Oxidase: Identification of the Two Cysteine Disulfide Bonds and Crystal Structure of A Non-Convertible Rat Liver Xanthine Dehydrogenase Mutant J.Biol.Chem. V. 280 24888 2005.
ISSN: ISSN 0021-9258
PubMed: 15878860
DOI: 10.1074/JBC.M501830200
Page generated: Tue Jul 8 03:22:14 2025
ISSN: ISSN 0021-9258
PubMed: 15878860
DOI: 10.1074/JBC.M501830200
Last articles
Cl in 8EBKCl in 8EA5
Cl in 8E9Y
Cl in 8EAD
Cl in 8EB3
Cl in 8E6A
Cl in 8E6M
Cl in 8E61
Cl in 8E6L
Cl in 8E6I