Calcium »
PDB 2i5w-2imw »
2iav »
Calcium in PDB 2iav: Crystal Structure of Squid Ganglion Dfpase H287A Mutant
Enzymatic activity of Crystal Structure of Squid Ganglion Dfpase H287A Mutant
All present enzymatic activity of Crystal Structure of Squid Ganglion Dfpase H287A Mutant:
3.1.8.2;
3.1.8.2;
Protein crystallography data
The structure of Crystal Structure of Squid Ganglion Dfpase H287A Mutant, PDB code: 2iav
was solved by
V.Katsemi,
C.Luecke,
J.Koepke,
F.Loehr,
S.Maurer,
G.Fritzsch,
H.Rueterjans,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 59.76 / 1.07 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 43.070, 81.996, 86.627, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.2 / 19.6 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of Squid Ganglion Dfpase H287A Mutant
(pdb code 2iav). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Crystal Structure of Squid Ganglion Dfpase H287A Mutant, PDB code: 2iav:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Crystal Structure of Squid Ganglion Dfpase H287A Mutant, PDB code: 2iav:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 2iav
Go back to
Calcium binding site 1 out
of 2 in the Crystal Structure of Squid Ganglion Dfpase H287A Mutant
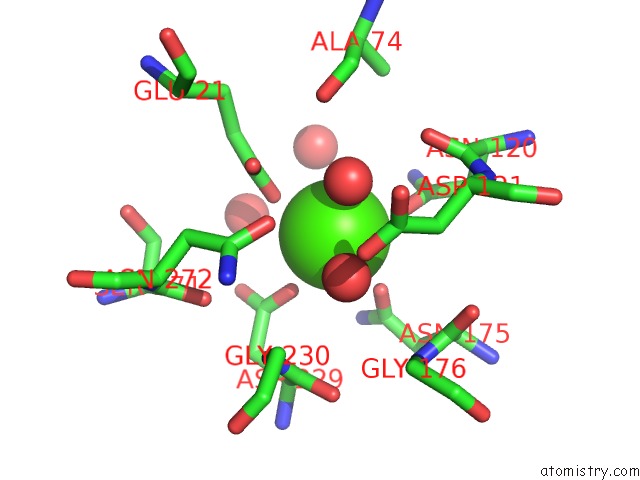
Mono view
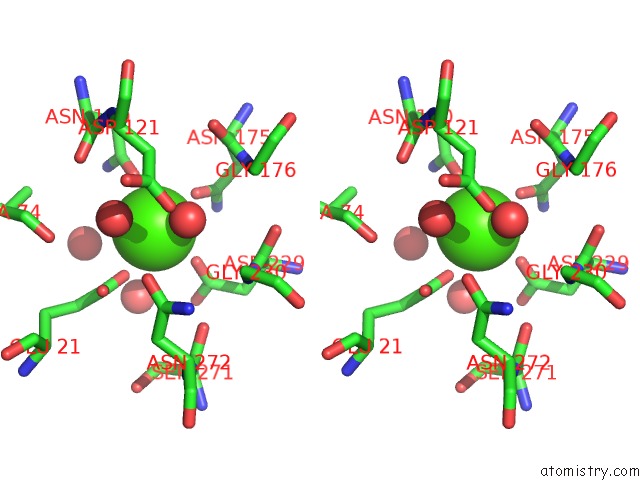
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of Squid Ganglion Dfpase H287A Mutant within 5.0Å range:
|
Calcium binding site 2 out of 2 in 2iav
Go back to
Calcium binding site 2 out
of 2 in the Crystal Structure of Squid Ganglion Dfpase H287A Mutant
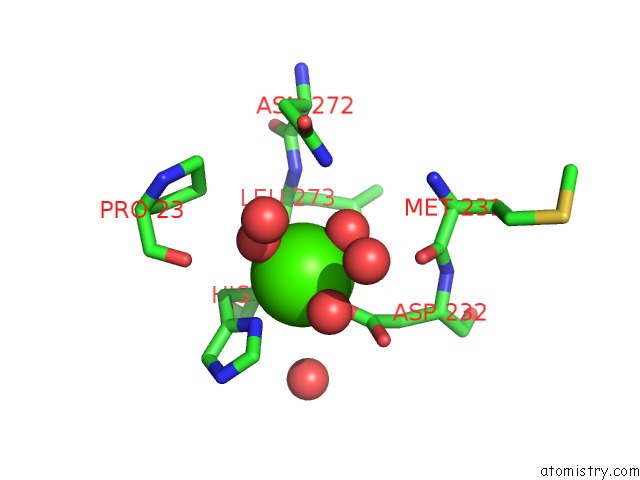
Mono view
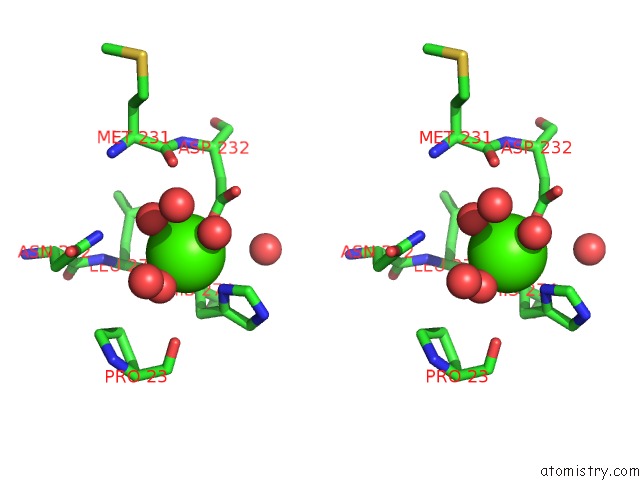
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Crystal Structure of Squid Ganglion Dfpase H287A Mutant within 5.0Å range:
|
Reference:
V.Katsemi,
C.Luecke,
J.Koepke,
F.Loehr,
S.Maurer,
G.Fritzsch,
H.Rueterjans.
Mutational and Structural Studies of the Diisopropylfluorophosphatase From Loligo Vulgaris Shed New Light on the Catalytic Mechanism of the Enzyme Biochemistry V. 44 9022 2005.
ISSN: ISSN 0006-2960
PubMed: 15966726
DOI: 10.1021/BI0500675
Page generated: Tue Jul 8 06:12:17 2025
ISSN: ISSN 0006-2960
PubMed: 15966726
DOI: 10.1021/BI0500675
Last articles
Cl in 8CX8Cl in 8CYU
Cl in 8CWJ
Cl in 8CXJ
Cl in 8CXL
Cl in 8CWH
Cl in 8CX9
Cl in 8CWI
Cl in 8CX5
Cl in 8CWG