Calcium »
PDB 2jdk-2jnx »
2jm4 »
Calcium in PDB 2jm4: The Solution uc(Nmr) Structure of the Relaxin (RXFP1) Receptor Ldla Module.
Calcium Binding Sites:
The binding sites of Calcium atom in the The Solution uc(Nmr) Structure of the Relaxin (RXFP1) Receptor Ldla Module.
(pdb code 2jm4). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total only one binding site of Calcium was determined in the The Solution uc(Nmr) Structure of the Relaxin (RXFP1) Receptor Ldla Module., PDB code: 2jm4:
In total only one binding site of Calcium was determined in the The Solution uc(Nmr) Structure of the Relaxin (RXFP1) Receptor Ldla Module., PDB code: 2jm4:
Calcium binding site 1 out of 1 in 2jm4
Go back to
Calcium binding site 1 out
of 1 in the The Solution uc(Nmr) Structure of the Relaxin (RXFP1) Receptor Ldla Module.
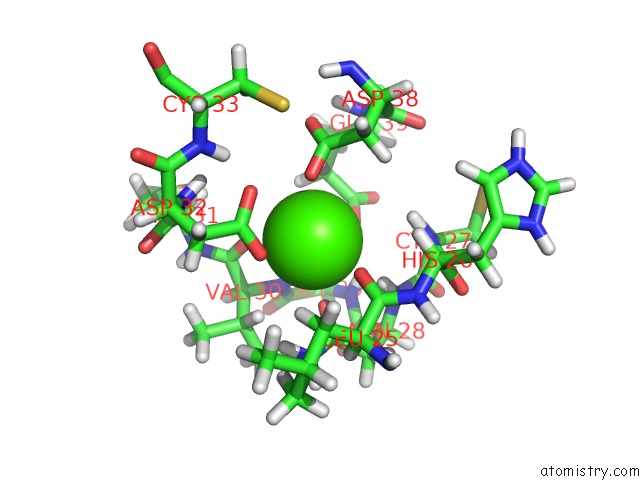
Mono view
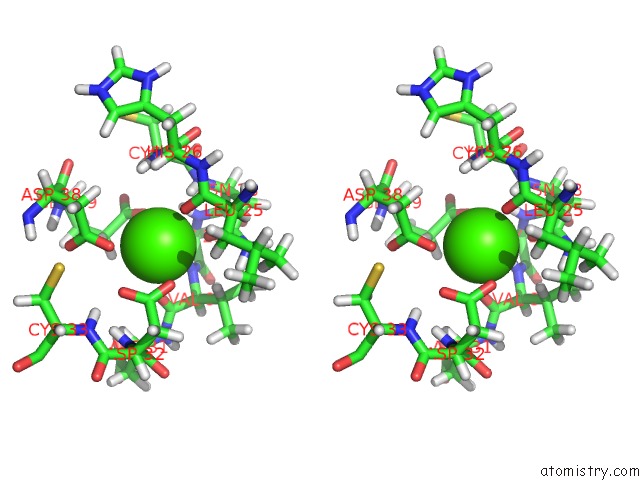
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of The Solution uc(Nmr) Structure of the Relaxin (RXFP1) Receptor Ldla Module. within 5.0Å range:
|
Reference:
E.J.Hopkins,
S.Layfield,
T.Ferraro,
R.A.D.Bathgate,
P.R.Gooley.
The uc(Nmr) Solution Structure of the Relaxin (RXFP1) Receptor Lipoprotein Receptor Class A Module and Identification of Key Residues in the N-Terminal Region of the Module That Mediate Receptor Activation J.Biol.Chem. V. 282 4172 2007.
ISSN: ISSN 0021-9258
PubMed: 17148455
DOI: 10.1074/JBC.M609526200
Page generated: Fri Jul 12 13:48:28 2024
ISSN: ISSN 0021-9258
PubMed: 17148455
DOI: 10.1074/JBC.M609526200
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1