Calcium »
PDB 2kug-2m0j »
2l1w »
Calcium in PDB 2l1w: The Solution Structure of Soybean Calmodulin Isoform 4 Complexed with the Vacuolar Calcium Atpase BCA1 Peptide
Calcium Binding Sites:
The binding sites of Calcium atom in the The Solution Structure of Soybean Calmodulin Isoform 4 Complexed with the Vacuolar Calcium Atpase BCA1 Peptide
(pdb code 2l1w). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 4 binding sites of Calcium where determined in the The Solution Structure of Soybean Calmodulin Isoform 4 Complexed with the Vacuolar Calcium Atpase BCA1 Peptide, PDB code: 2l1w:
Jump to Calcium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Calcium where determined in the The Solution Structure of Soybean Calmodulin Isoform 4 Complexed with the Vacuolar Calcium Atpase BCA1 Peptide, PDB code: 2l1w:
Jump to Calcium binding site number: 1; 2; 3; 4;
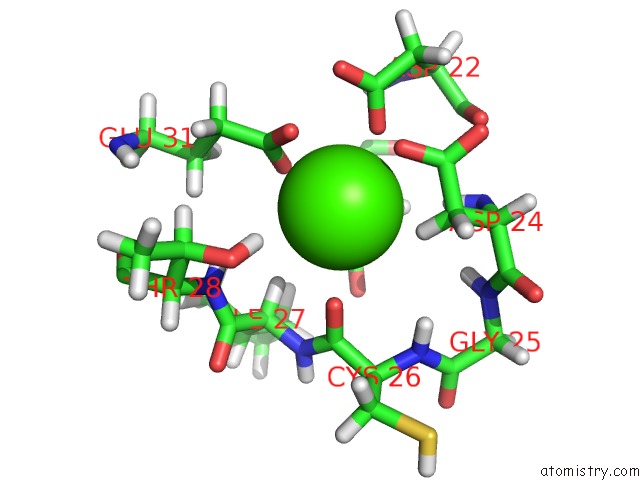
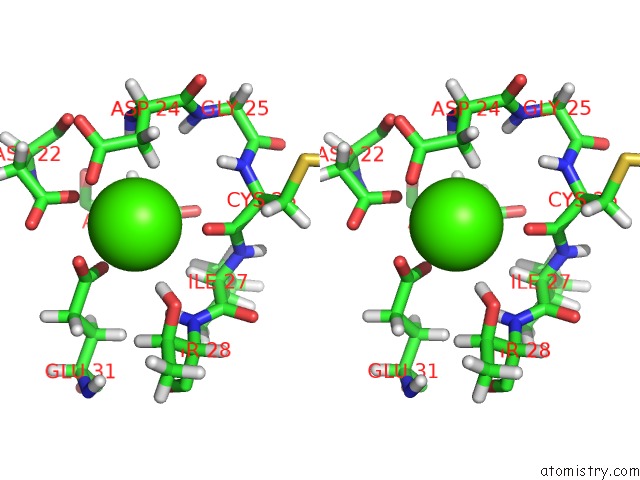
Calcium binding site 1 out of 4 in 2l1w
Go back to
Calcium binding site 1 out
of 4 in the The Solution Structure of Soybean Calmodulin Isoform 4 Complexed with the Vacuolar Calcium Atpase BCA1 Peptide
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of The Solution Structure of Soybean Calmodulin Isoform 4 Complexed with the Vacuolar Calcium Atpase BCA1 Peptide within 5.0Å range:
|
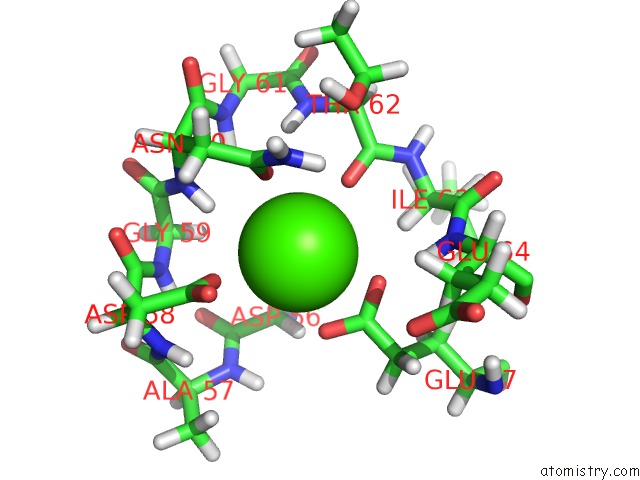
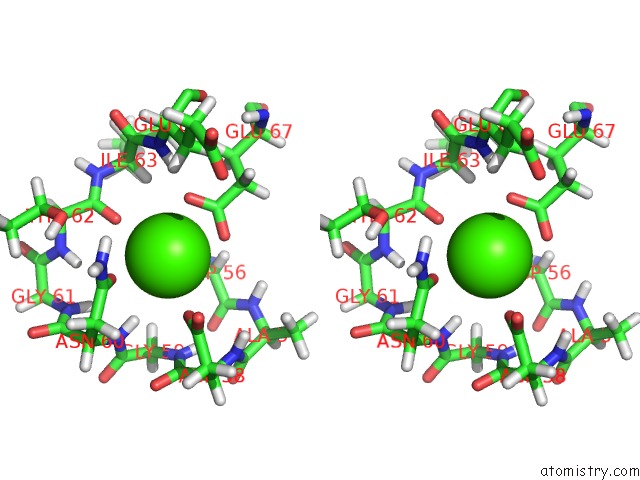
Calcium binding site 2 out of 4 in 2l1w
Go back to
Calcium binding site 2 out
of 4 in the The Solution Structure of Soybean Calmodulin Isoform 4 Complexed with the Vacuolar Calcium Atpase BCA1 Peptide
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of The Solution Structure of Soybean Calmodulin Isoform 4 Complexed with the Vacuolar Calcium Atpase BCA1 Peptide within 5.0Å range:
|
Calcium binding site 3 out of 4 in 2l1w
Go back to
Calcium binding site 3 out
of 4 in the The Solution Structure of Soybean Calmodulin Isoform 4 Complexed with the Vacuolar Calcium Atpase BCA1 Peptide
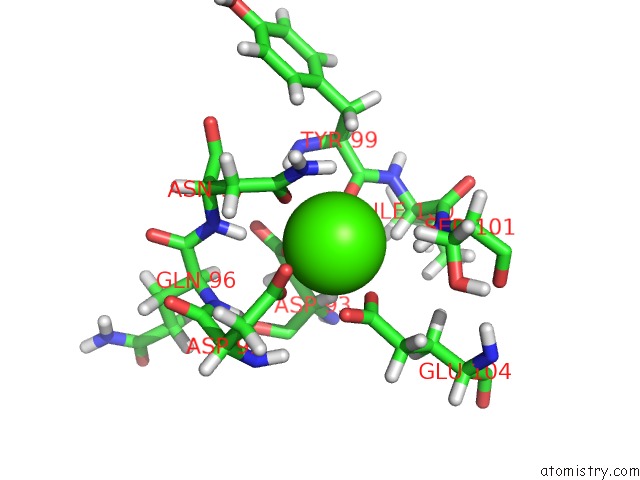
Mono view
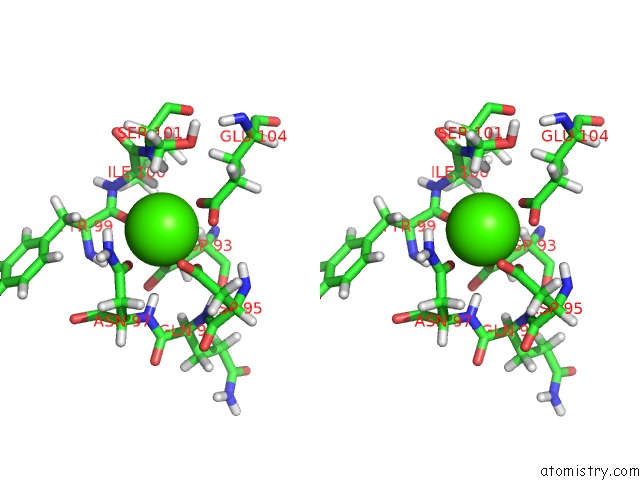
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of The Solution Structure of Soybean Calmodulin Isoform 4 Complexed with the Vacuolar Calcium Atpase BCA1 Peptide within 5.0Å range:
|
Calcium binding site 4 out of 4 in 2l1w
Go back to
Calcium binding site 4 out
of 4 in the The Solution Structure of Soybean Calmodulin Isoform 4 Complexed with the Vacuolar Calcium Atpase BCA1 Peptide
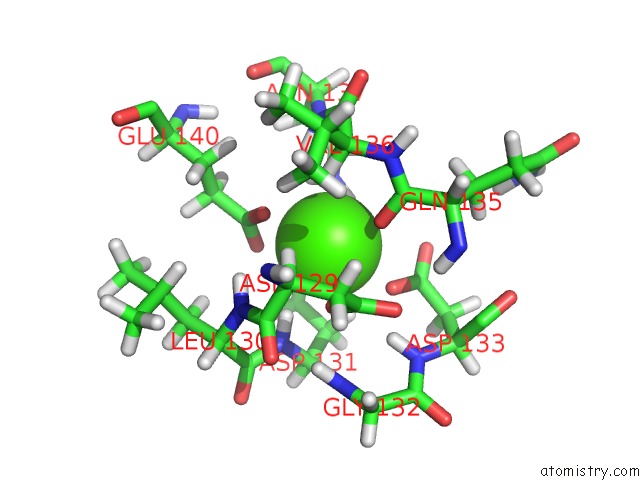
Mono view
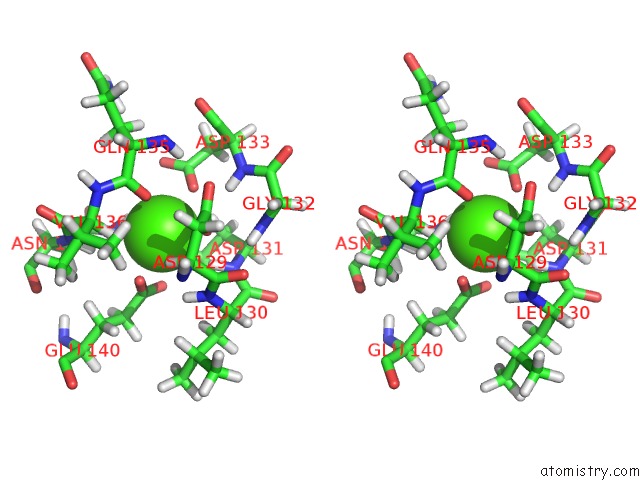
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of The Solution Structure of Soybean Calmodulin Isoform 4 Complexed with the Vacuolar Calcium Atpase BCA1 Peptide within 5.0Å range:
|
Reference:
H.Ishida,
H.Vogel.
The Solution Structure of A Plant Calmodulin and the Cam-Binding Domain of the Vacuolar Calcium-Atpase BCA1 Reveals A New Binding and Activation Mechanism To Be Published.
Page generated: Tue Jul 8 06:54:35 2025
Last articles
Ca in 3FU1Ca in 3FSJ
Ca in 3FRP
Ca in 3FPK
Ca in 3FP8
Ca in 3FP7
Ca in 3FP6
Ca in 3FO3
Ca in 3FOR
Ca in 3FOU