Calcium »
PDB 2kug-2m0j »
2lp3 »
Calcium in PDB 2lp3: Solution Structure of S100A1 CA2+
Calcium Binding Sites:
The binding sites of Calcium atom in the Solution Structure of S100A1 CA2+
(pdb code 2lp3). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 4 binding sites of Calcium where determined in the Solution Structure of S100A1 CA2+, PDB code: 2lp3:
Jump to Calcium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Calcium where determined in the Solution Structure of S100A1 CA2+, PDB code: 2lp3:
Jump to Calcium binding site number: 1; 2; 3; 4;
Calcium binding site 1 out of 4 in 2lp3
Go back to
Calcium binding site 1 out
of 4 in the Solution Structure of S100A1 CA2+
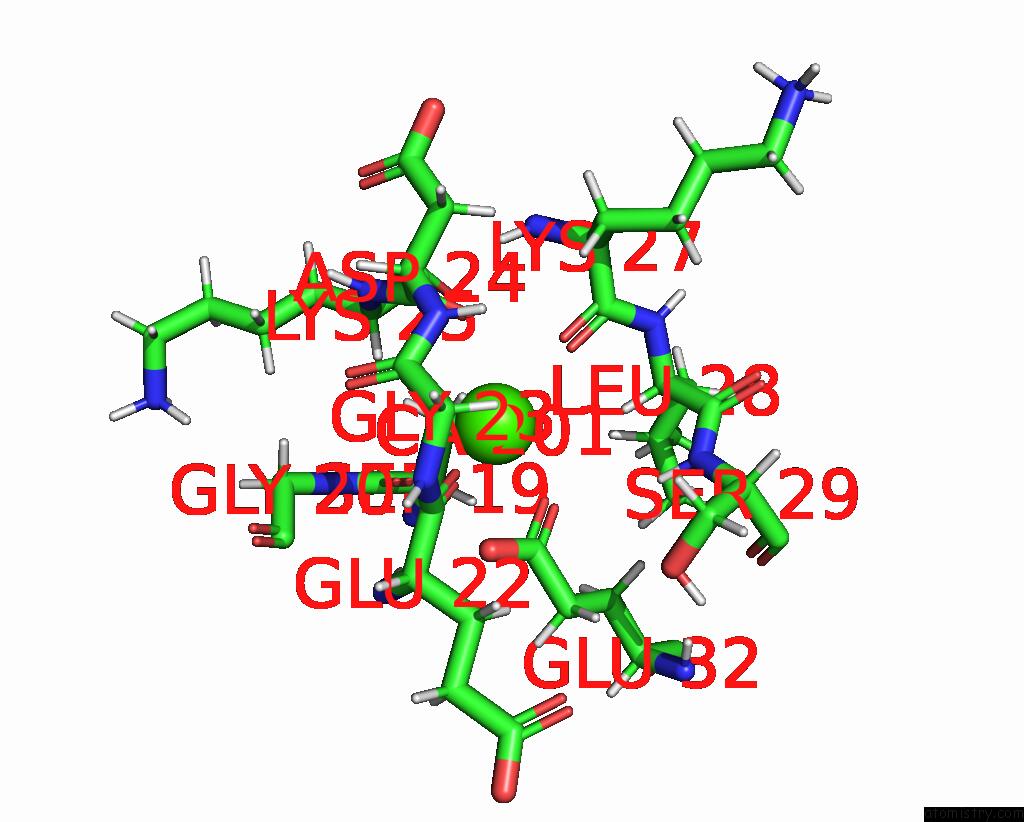
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Solution Structure of S100A1 CA2+ within 5.0Å range:
|
Calcium binding site 2 out of 4 in 2lp3
Go back to
Calcium binding site 2 out
of 4 in the Solution Structure of S100A1 CA2+

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Solution Structure of S100A1 CA2+ within 5.0Å range:
|
Calcium binding site 3 out of 4 in 2lp3
Go back to
Calcium binding site 3 out
of 4 in the Solution Structure of S100A1 CA2+

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Solution Structure of S100A1 CA2+ within 5.0Å range:
|
Calcium binding site 4 out of 4 in 2lp3
Go back to
Calcium binding site 4 out
of 4 in the Solution Structure of S100A1 CA2+

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of Solution Structure of S100A1 CA2+ within 5.0Å range:
|
Reference:
M.Nowakowski,
K.Ruszczynska-Bartnik,
M.Budzinska,
L.Jaremko,
M.Jaremko,
K.Zdanowski,
A.Bierzynski,
A.Ejchart.
Impact of Calcium Binding and Thionylation of S100A1 Protein on Its Nuclear Magnetic Resonance-Derived Structure and Backbone Dynamics. Biochemistry V. 52 1149 2013.
ISSN: ISSN 0006-2960
PubMed: 23351007
DOI: 10.1021/BI3015407
Page generated: Tue Jul 8 06:59:30 2025
ISSN: ISSN 0006-2960
PubMed: 23351007
DOI: 10.1021/BI3015407
Last articles
Ca in 3E3YCa in 3DVO
Ca in 3E0D
Ca in 3E11
Ca in 3DZM
Ca in 3E03
Ca in 3DZZ
Ca in 3DYB
Ca in 3DZC
Ca in 3DXP