Calcium »
PDB 2kug-2m0j »
2lqp »
Calcium in PDB 2lqp: uc(Nmr) Solution Structure of the CA2+-Calmodulin C-Terminal Domain in A Complex with A Peptide (Nscate) From the L-Type Voltage-Gated Calcium Channel ALPHA1C Subunit
Calcium Binding Sites:
The binding sites of Calcium atom in the uc(Nmr) Solution Structure of the CA2+-Calmodulin C-Terminal Domain in A Complex with A Peptide (Nscate) From the L-Type Voltage-Gated Calcium Channel ALPHA1C Subunit
(pdb code 2lqp). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the uc(Nmr) Solution Structure of the CA2+-Calmodulin C-Terminal Domain in A Complex with A Peptide (Nscate) From the L-Type Voltage-Gated Calcium Channel ALPHA1C Subunit, PDB code: 2lqp:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the uc(Nmr) Solution Structure of the CA2+-Calmodulin C-Terminal Domain in A Complex with A Peptide (Nscate) From the L-Type Voltage-Gated Calcium Channel ALPHA1C Subunit, PDB code: 2lqp:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 2lqp
Go back to
Calcium binding site 1 out
of 2 in the uc(Nmr) Solution Structure of the CA2+-Calmodulin C-Terminal Domain in A Complex with A Peptide (Nscate) From the L-Type Voltage-Gated Calcium Channel ALPHA1C Subunit
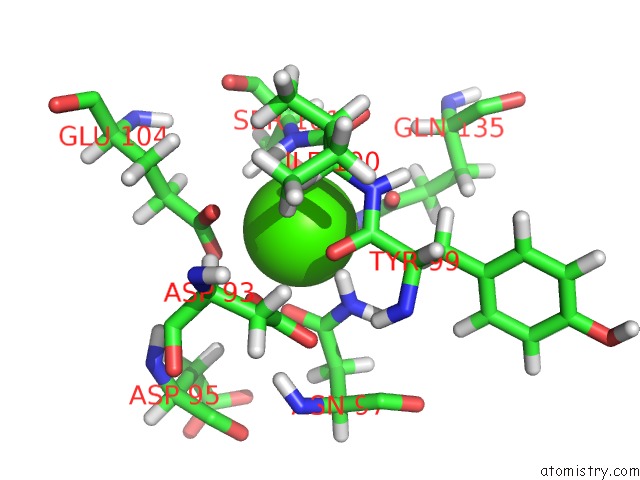
Mono view
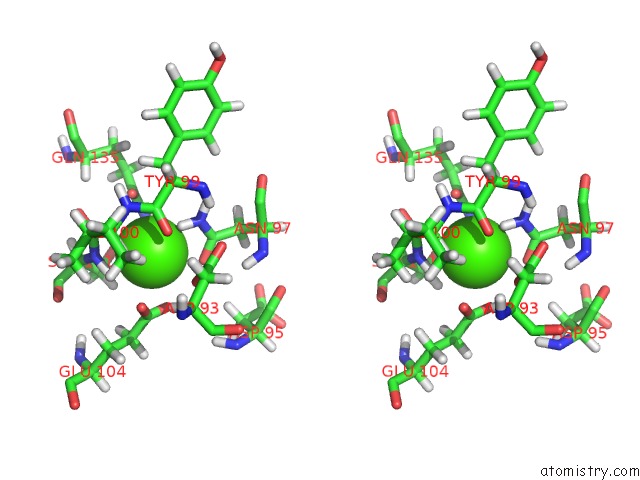
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of uc(Nmr) Solution Structure of the CA2+-Calmodulin C-Terminal Domain in A Complex with A Peptide (Nscate) From the L-Type Voltage-Gated Calcium Channel ALPHA1C Subunit within 5.0Å range:
|
Calcium binding site 2 out of 2 in 2lqp
Go back to
Calcium binding site 2 out
of 2 in the uc(Nmr) Solution Structure of the CA2+-Calmodulin C-Terminal Domain in A Complex with A Peptide (Nscate) From the L-Type Voltage-Gated Calcium Channel ALPHA1C Subunit
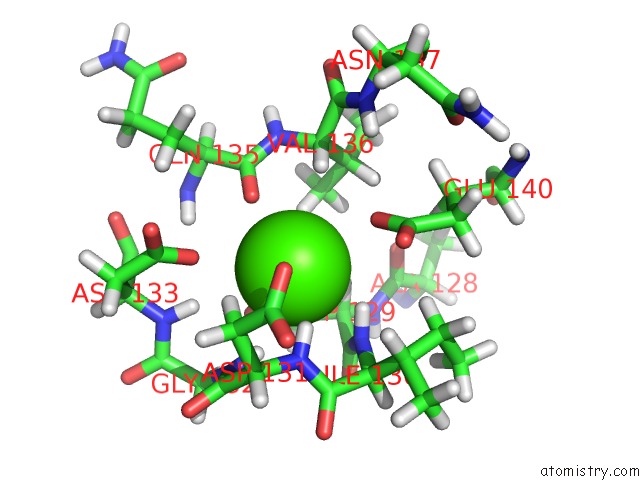
Mono view
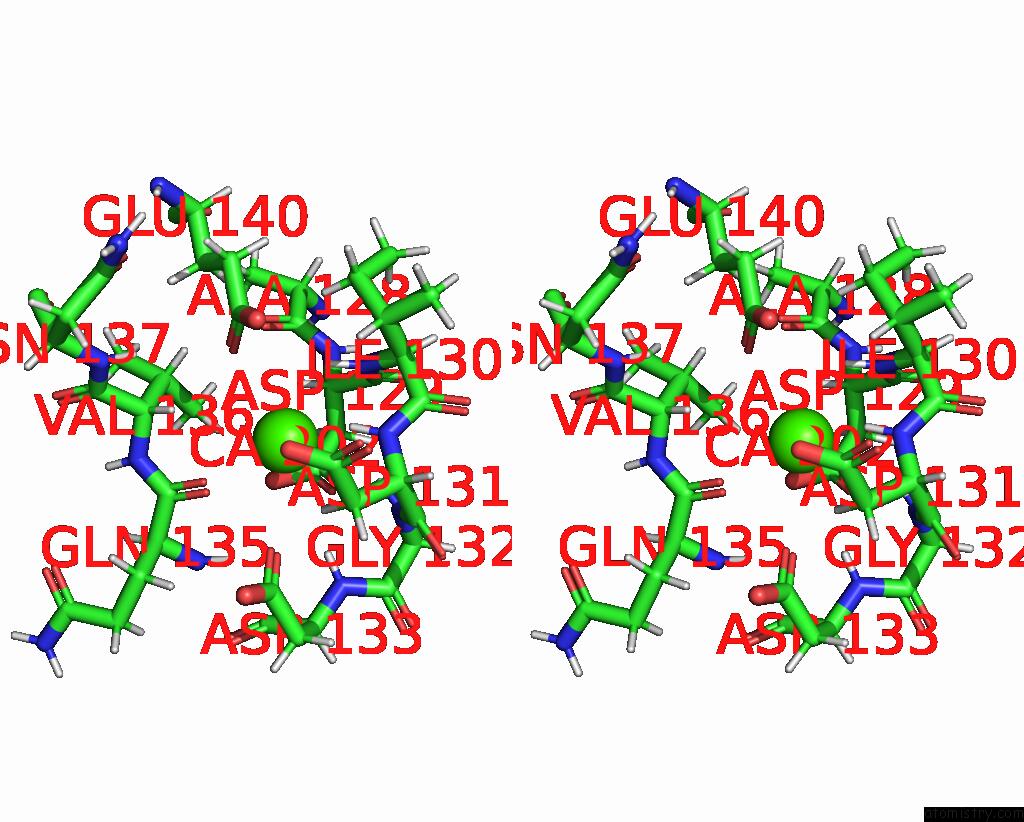
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of uc(Nmr) Solution Structure of the CA2+-Calmodulin C-Terminal Domain in A Complex with A Peptide (Nscate) From the L-Type Voltage-Gated Calcium Channel ALPHA1C Subunit within 5.0Å range:
|
Reference:
Z.Liu,
H.J.Vogel.
Structural Basis For the Regulation of L-Type Voltage-Gated Calcium Channels: Interactions Between the N-Terminal Cytoplasmic Domain and Ca(2+)-Calmodulin. Front Mol Neurosci V. 5 38 2012.
ISSN: ESSN 1662-5099
PubMed: 22518098
DOI: 10.3389/FNMOL.2012.00038
Page generated: Tue Jul 8 06:59:51 2025
ISSN: ESSN 1662-5099
PubMed: 22518098
DOI: 10.3389/FNMOL.2012.00038
Last articles
Ca in 3IJECa in 3IK2
Ca in 3IJ9
Ca in 3IJ8
Ca in 3IJ7
Ca in 3IGO
Ca in 3IIT
Ca in 3IFK
Ca in 3IAS
Ca in 3I9H