Calcium »
PDB 2xc5-2xon »
2xfg »
Calcium in PDB 2xfg: Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules
Enzymatic activity of Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules
All present enzymatic activity of Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules:
3.2.1.4;
3.2.1.4;
Protein crystallography data
The structure of Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules, PDB code: 2xfg
was solved by
S.Petkun,
R.Lamed,
S.Jindou,
T.Burstein,
O.Yaniv,
Y.Shoham,
J.W.L.Shimon,
E.A.Bayer,
F.Frolow,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.868 / 1.68 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 70.399, 88.541, 106.486, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.14 / 17.59 |
Other elements in 2xfg:
The structure of Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules also contains other interesting chemical elements:
| Chlorine | (Cl) | 3 atoms |
Calcium Binding Sites:
The binding sites of Calcium atom in the Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules
(pdb code 2xfg). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules, PDB code: 2xfg:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules, PDB code: 2xfg:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 2xfg
Go back to
Calcium binding site 1 out
of 2 in the Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules within 5.0Å range:
|
Calcium binding site 2 out of 2 in 2xfg
Go back to
Calcium binding site 2 out
of 2 in the Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules

Mono view
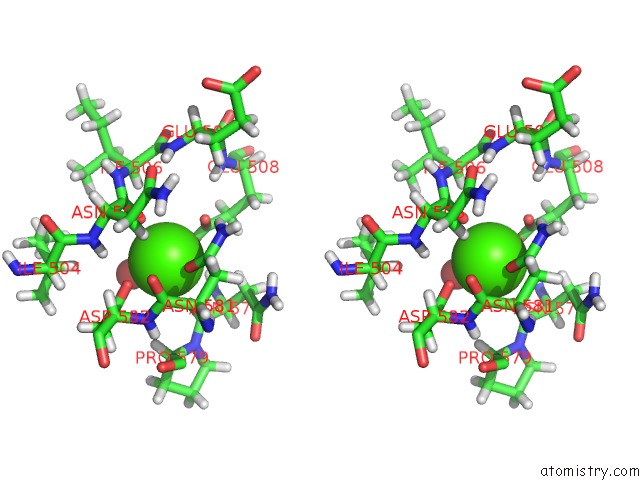
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules within 5.0Å range:
|
Reference:
S.Petkun,
I.R.Grinberg,
R.Lamed,
S.Jindou,
T.Burstein,
O.Yaniv,
Y.Shoham,
J.W.L.Shimon,
E.A.Bayer,
F.Frolow.
Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Its Component Parts: Structural and Functional Significance of Intermodular Linker Peerj V. 3 E1126 2015.
ISSN: ISSN 2167-8359
PubMed: 26401442
DOI: 10.7717/PEERJ.1126
Page generated: Tue Jul 8 09:21:50 2025
ISSN: ISSN 2167-8359
PubMed: 26401442
DOI: 10.7717/PEERJ.1126
Last articles
F in 4I9OF in 4IA9
F in 4I89
F in 4I7S
F in 4I87
F in 4I8N
F in 4I7Q
F in 4I85
F in 4I1O
F in 4HY0