Calcium »
PDB 2xqg-2y6h »
2xy8 »
Calcium in PDB 2xy8: Paramagnetic-Based uc(Nmr) Structure of the Complex Between the N- Terminal Epsilon Domain and the Theta Domain of the Dna Polymerase III
Enzymatic activity of Paramagnetic-Based uc(Nmr) Structure of the Complex Between the N- Terminal Epsilon Domain and the Theta Domain of the Dna Polymerase III
All present enzymatic activity of Paramagnetic-Based uc(Nmr) Structure of the Complex Between the N- Terminal Epsilon Domain and the Theta Domain of the Dna Polymerase III:
2.7.7.7;
2.7.7.7;
Calcium Binding Sites:
The binding sites of Calcium atom in the Paramagnetic-Based uc(Nmr) Structure of the Complex Between the N- Terminal Epsilon Domain and the Theta Domain of the Dna Polymerase III
(pdb code 2xy8). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total only one binding site of Calcium was determined in the Paramagnetic-Based uc(Nmr) Structure of the Complex Between the N- Terminal Epsilon Domain and the Theta Domain of the Dna Polymerase III, PDB code: 2xy8:
In total only one binding site of Calcium was determined in the Paramagnetic-Based uc(Nmr) Structure of the Complex Between the N- Terminal Epsilon Domain and the Theta Domain of the Dna Polymerase III, PDB code: 2xy8:
Calcium binding site 1 out of 1 in 2xy8
Go back to
Calcium binding site 1 out
of 1 in the Paramagnetic-Based uc(Nmr) Structure of the Complex Between the N- Terminal Epsilon Domain and the Theta Domain of the Dna Polymerase III
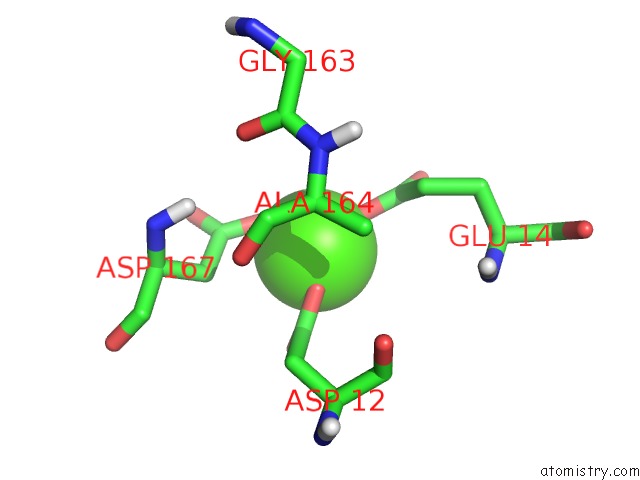
Mono view
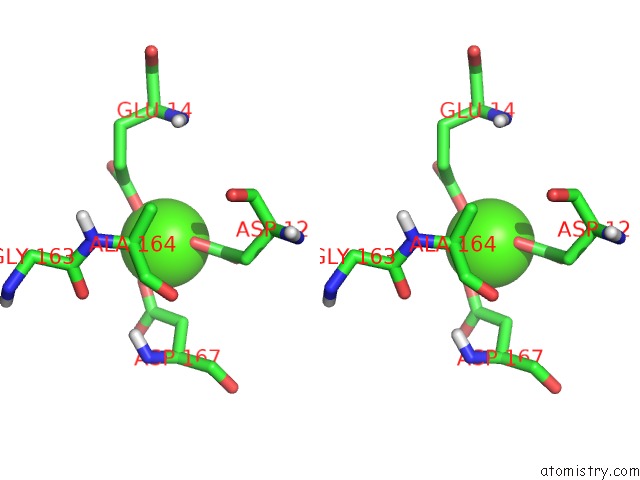
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Paramagnetic-Based uc(Nmr) Structure of the Complex Between the N- Terminal Epsilon Domain and the Theta Domain of the Dna Polymerase III within 5.0Å range:
|
Reference:
C.Schmitz,
A.M.J.J.Bonvin.
Protein-Protein Haddocking Using Exclusively Pseudocontact Shifts. J.Biomol.uc(Nmr) V. 50 263 2011.
ISSN: ISSN 0925-2738
PubMed: 21626213
DOI: 10.1007/S10858-011-9514-4
Page generated: Fri Jul 12 19:17:14 2024
ISSN: ISSN 0925-2738
PubMed: 21626213
DOI: 10.1007/S10858-011-9514-4
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1