Calcium »
PDB 3iti-3k3w »
3ju9 »
Calcium in PDB 3ju9: Crystal Structure of A Lectin From Canavalia Brasiliensis Seed (Conbr) Complexed with Alpha-Aminobutyric Acid
Protein crystallography data
The structure of Crystal Structure of A Lectin From Canavalia Brasiliensis Seed (Conbr) Complexed with Alpha-Aminobutyric Acid, PDB code: 3ju9
was solved by
E.H.S.Bezerra,
B.A.M.Rocha,
C.S.Nagano,
G.A.Bezerra,
T.R.Moura,
M.J.B.Bezerra,
R.G.Benevides,
E.S.Marinho,
P.Delatorre,
B.S.Cavada,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 36.51 / 2.10 |
| Space group | I 2 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 68.323, 73.020, 99.542, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.4 / 25.3 |
Other elements in 3ju9:
The structure of Crystal Structure of A Lectin From Canavalia Brasiliensis Seed (Conbr) Complexed with Alpha-Aminobutyric Acid also contains other interesting chemical elements:
| Manganese | (Mn) | 1 atom |
| Chlorine | (Cl) | 2 atoms |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of A Lectin From Canavalia Brasiliensis Seed (Conbr) Complexed with Alpha-Aminobutyric Acid
(pdb code 3ju9). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total only one binding site of Calcium was determined in the Crystal Structure of A Lectin From Canavalia Brasiliensis Seed (Conbr) Complexed with Alpha-Aminobutyric Acid, PDB code: 3ju9:
In total only one binding site of Calcium was determined in the Crystal Structure of A Lectin From Canavalia Brasiliensis Seed (Conbr) Complexed with Alpha-Aminobutyric Acid, PDB code: 3ju9:
Calcium binding site 1 out of 1 in 3ju9
Go back to
Calcium binding site 1 out
of 1 in the Crystal Structure of A Lectin From Canavalia Brasiliensis Seed (Conbr) Complexed with Alpha-Aminobutyric Acid
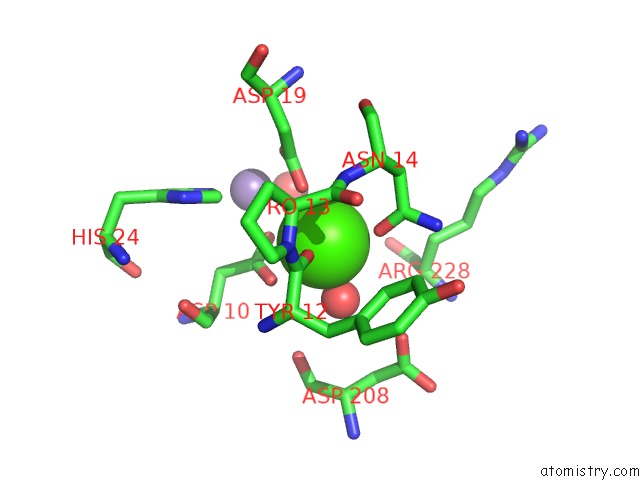
Mono view
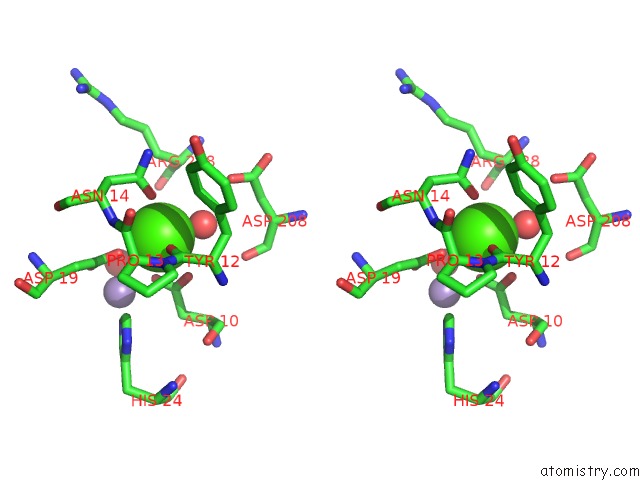
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of A Lectin From Canavalia Brasiliensis Seed (Conbr) Complexed with Alpha-Aminobutyric Acid within 5.0Å range:
|
Reference:
E.H.Bezerra,
B.A.Rocha,
C.S.Nagano,
G.A.Bezerra,
T.R.Moura,
M.J.Bezerra,
R.G.Benevides,
A.H.Sampaio,
A.M.Assreuy,
P.Delatorre,
B.S.Cavada.
Structural Analysis of Conbr Reveals Molecular Correlation Between the Carbohydrate Recognition Domain and Endothelial No Synthase Activation. Biochem.Biophys.Res.Commun. V. 408 566 2011.
ISSN: ISSN 0006-291X
PubMed: 21530490
DOI: 10.1016/J.BBRC.2011.04.061
Page generated: Tue Jul 8 13:33:54 2025
ISSN: ISSN 0006-291X
PubMed: 21530490
DOI: 10.1016/J.BBRC.2011.04.061
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO