Calcium »
PDB 3ngw-3nuc »
3nhg »
Calcium in PDB 3nhg: RB69 Dna Polymerase (S565G/Y567A) Ternary Complex with Dttp Opposite Dg
Enzymatic activity of RB69 Dna Polymerase (S565G/Y567A) Ternary Complex with Dttp Opposite Dg
All present enzymatic activity of RB69 Dna Polymerase (S565G/Y567A) Ternary Complex with Dttp Opposite Dg:
2.7.7.7;
2.7.7.7;
Protein crystallography data
The structure of RB69 Dna Polymerase (S565G/Y567A) Ternary Complex with Dttp Opposite Dg, PDB code: 3nhg
was solved by
M.Wang,
J.Wang,
W.H.Konigsberg,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.50 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 75.052, 119.999, 130.382, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.5 / 24.4 |
Calcium Binding Sites:
The binding sites of Calcium atom in the RB69 Dna Polymerase (S565G/Y567A) Ternary Complex with Dttp Opposite Dg
(pdb code 3nhg). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 6 binding sites of Calcium where determined in the RB69 Dna Polymerase (S565G/Y567A) Ternary Complex with Dttp Opposite Dg, PDB code: 3nhg:
Jump to Calcium binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Calcium where determined in the RB69 Dna Polymerase (S565G/Y567A) Ternary Complex with Dttp Opposite Dg, PDB code: 3nhg:
Jump to Calcium binding site number: 1; 2; 3; 4; 5; 6;
Calcium binding site 1 out of 6 in 3nhg
Go back to
Calcium binding site 1 out
of 6 in the RB69 Dna Polymerase (S565G/Y567A) Ternary Complex with Dttp Opposite Dg
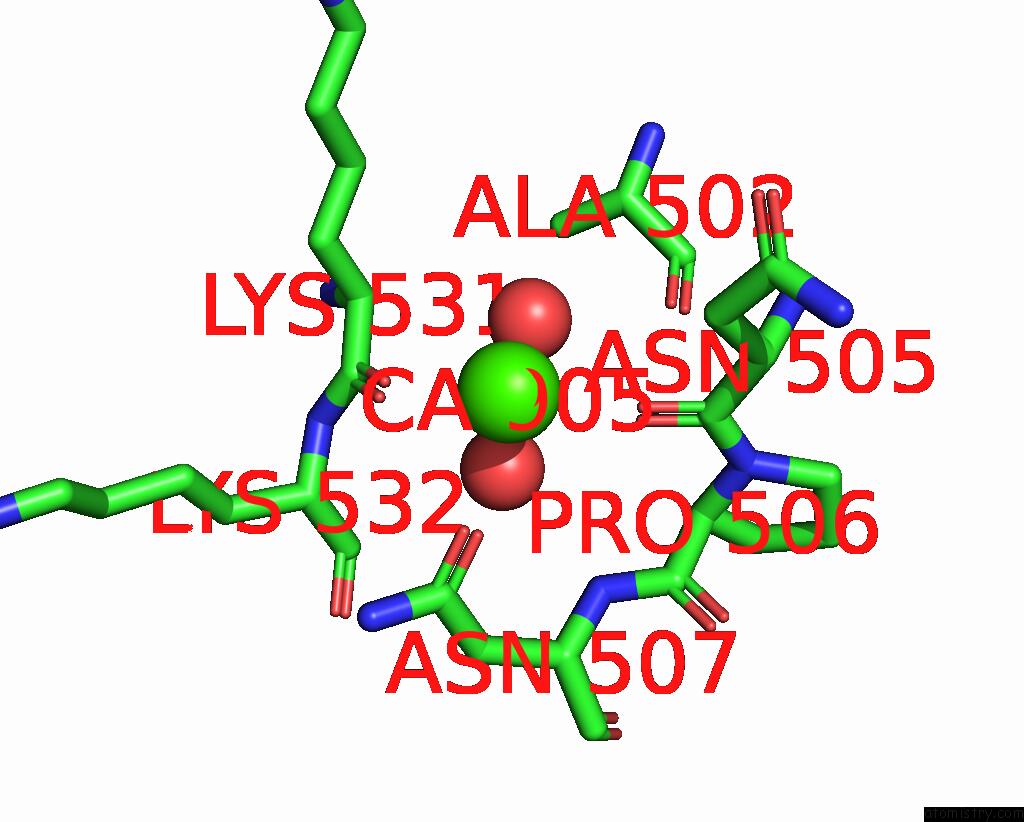
Mono view
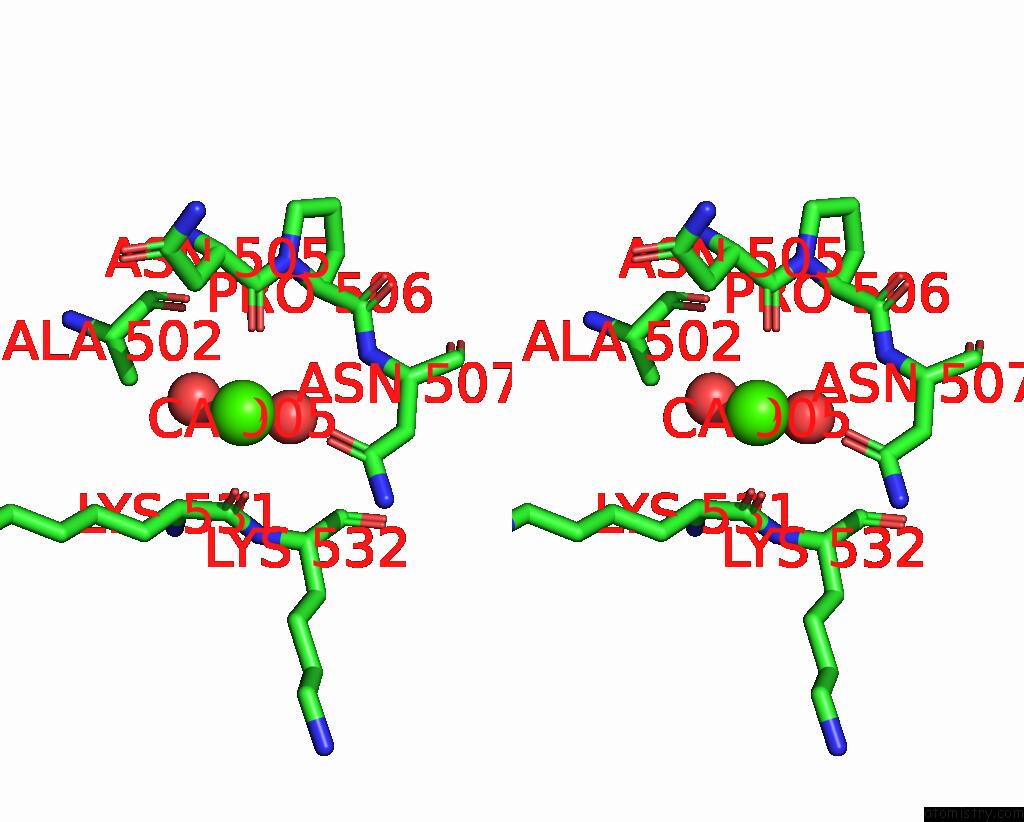
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of RB69 Dna Polymerase (S565G/Y567A) Ternary Complex with Dttp Opposite Dg within 5.0Å range:
|
Calcium binding site 2 out of 6 in 3nhg
Go back to
Calcium binding site 2 out
of 6 in the RB69 Dna Polymerase (S565G/Y567A) Ternary Complex with Dttp Opposite Dg
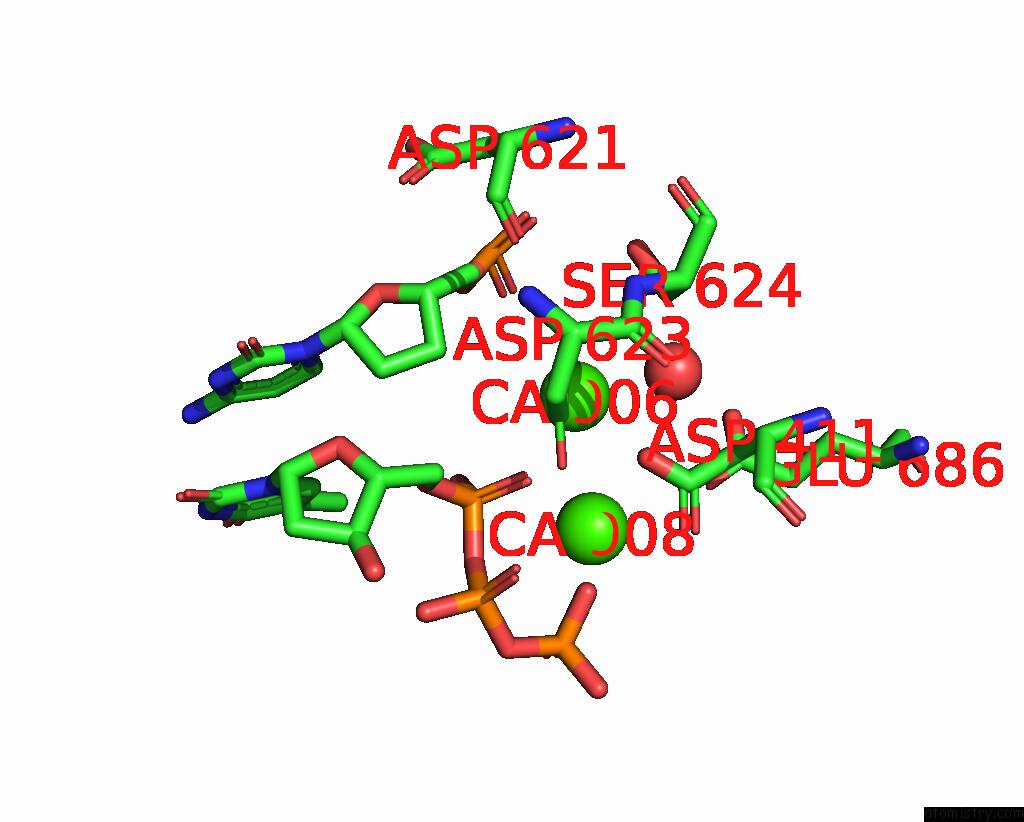
Mono view
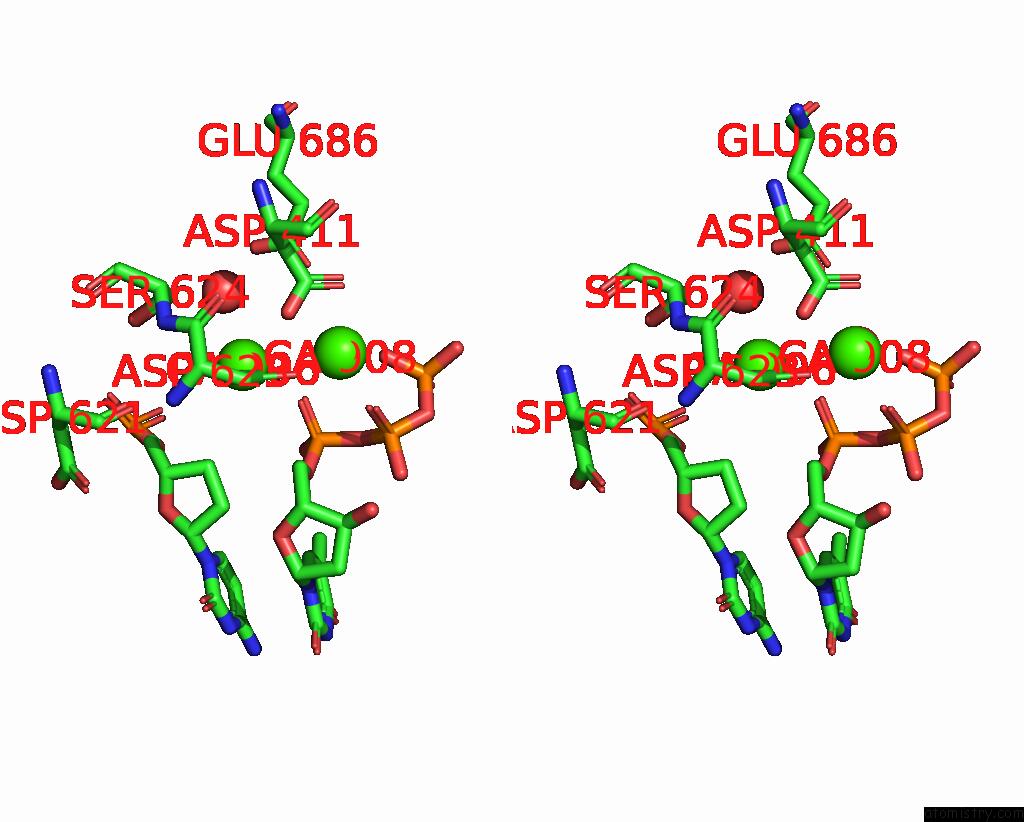
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of RB69 Dna Polymerase (S565G/Y567A) Ternary Complex with Dttp Opposite Dg within 5.0Å range:
|
Calcium binding site 3 out of 6 in 3nhg
Go back to
Calcium binding site 3 out
of 6 in the RB69 Dna Polymerase (S565G/Y567A) Ternary Complex with Dttp Opposite Dg
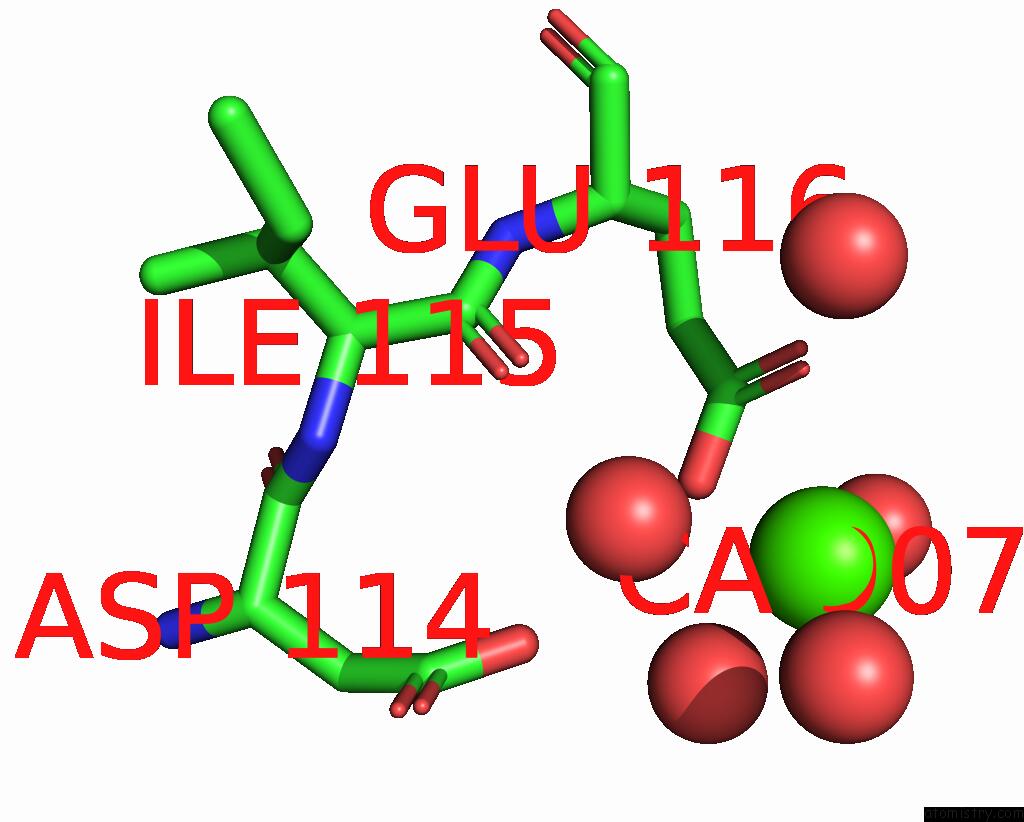
Mono view
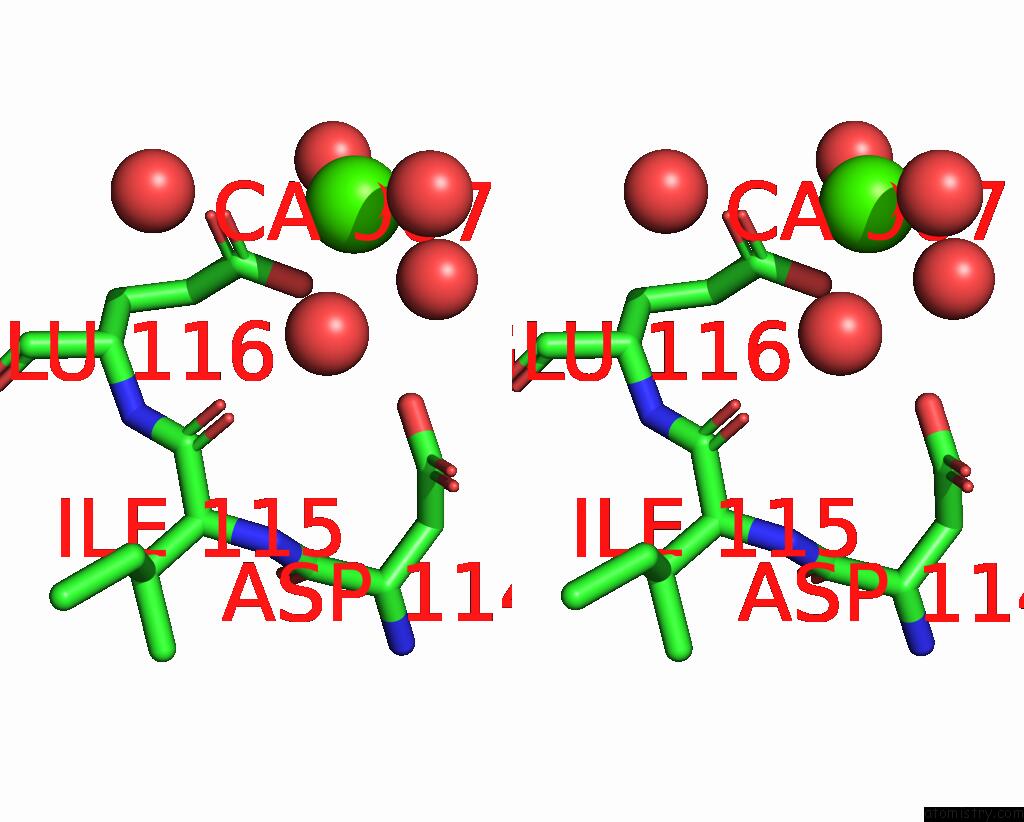
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of RB69 Dna Polymerase (S565G/Y567A) Ternary Complex with Dttp Opposite Dg within 5.0Å range:
|
Calcium binding site 4 out of 6 in 3nhg
Go back to
Calcium binding site 4 out
of 6 in the RB69 Dna Polymerase (S565G/Y567A) Ternary Complex with Dttp Opposite Dg
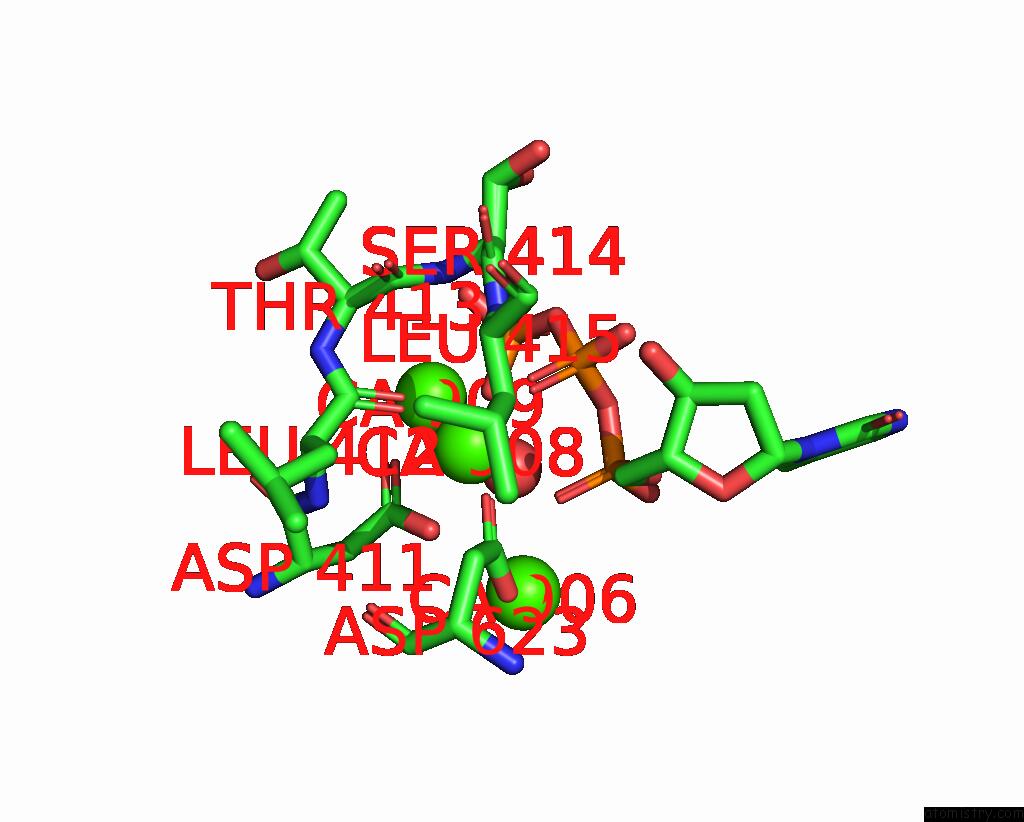
Mono view
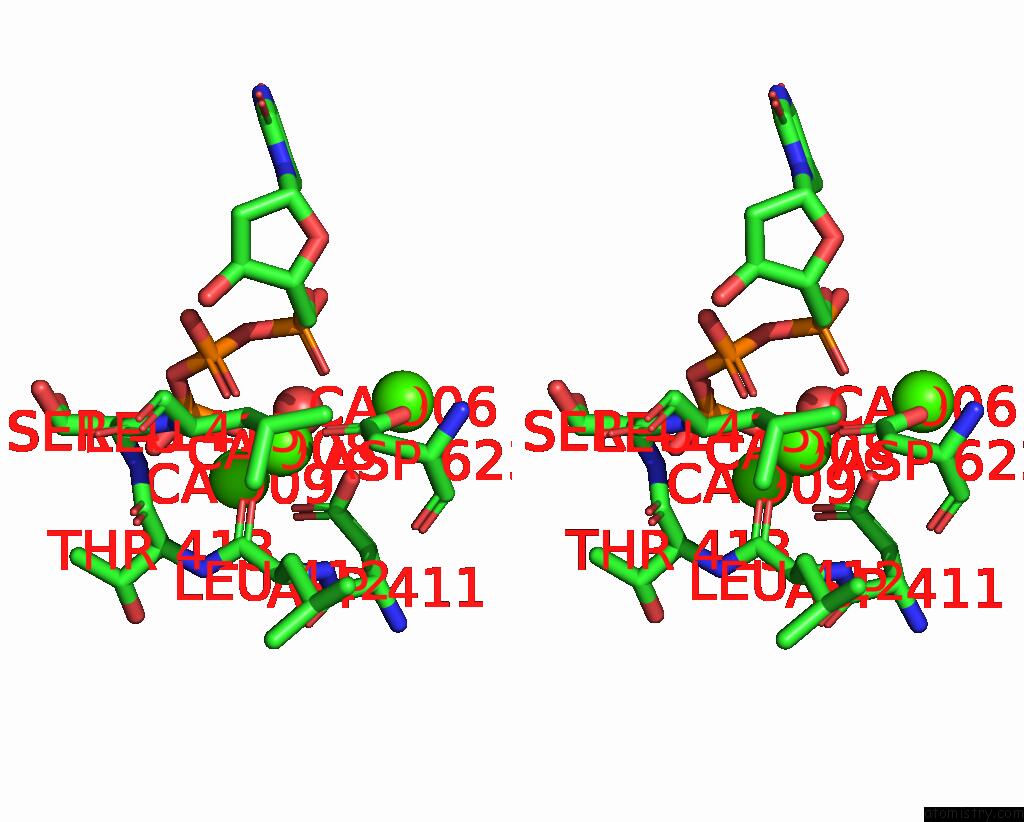
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of RB69 Dna Polymerase (S565G/Y567A) Ternary Complex with Dttp Opposite Dg within 5.0Å range:
|
Calcium binding site 5 out of 6 in 3nhg
Go back to
Calcium binding site 5 out
of 6 in the RB69 Dna Polymerase (S565G/Y567A) Ternary Complex with Dttp Opposite Dg
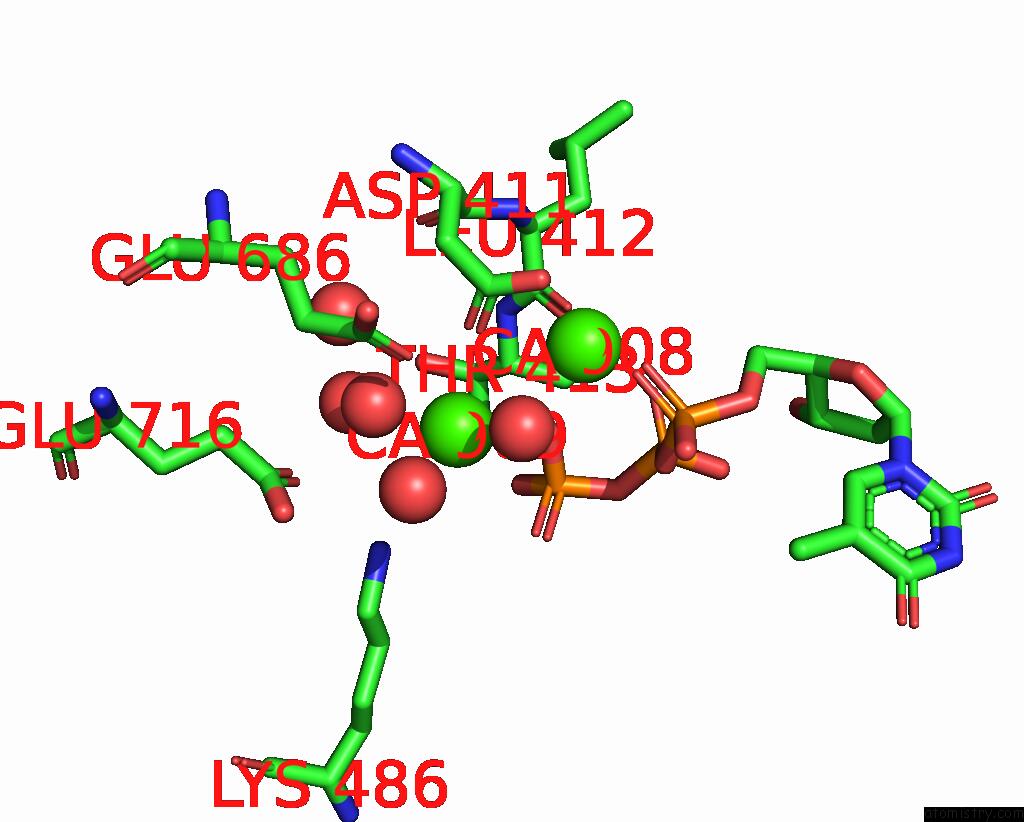
Mono view
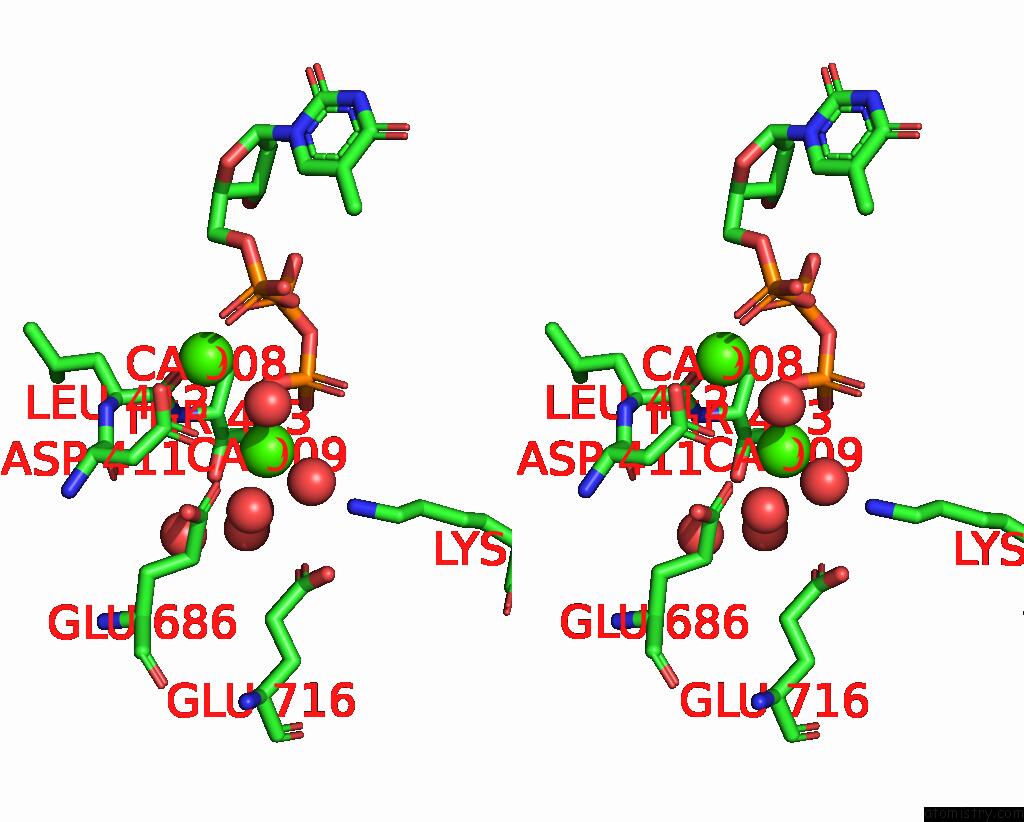
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 5 of RB69 Dna Polymerase (S565G/Y567A) Ternary Complex with Dttp Opposite Dg within 5.0Å range:
|
Calcium binding site 6 out of 6 in 3nhg
Go back to
Calcium binding site 6 out
of 6 in the RB69 Dna Polymerase (S565G/Y567A) Ternary Complex with Dttp Opposite Dg
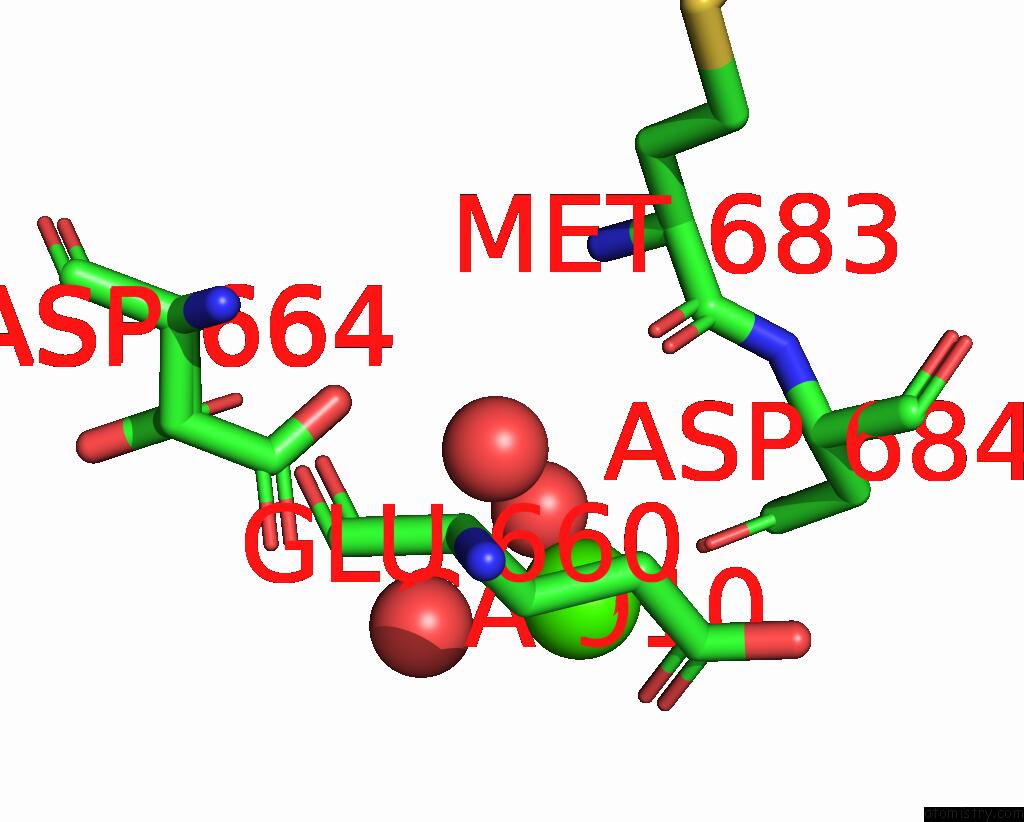
Mono view
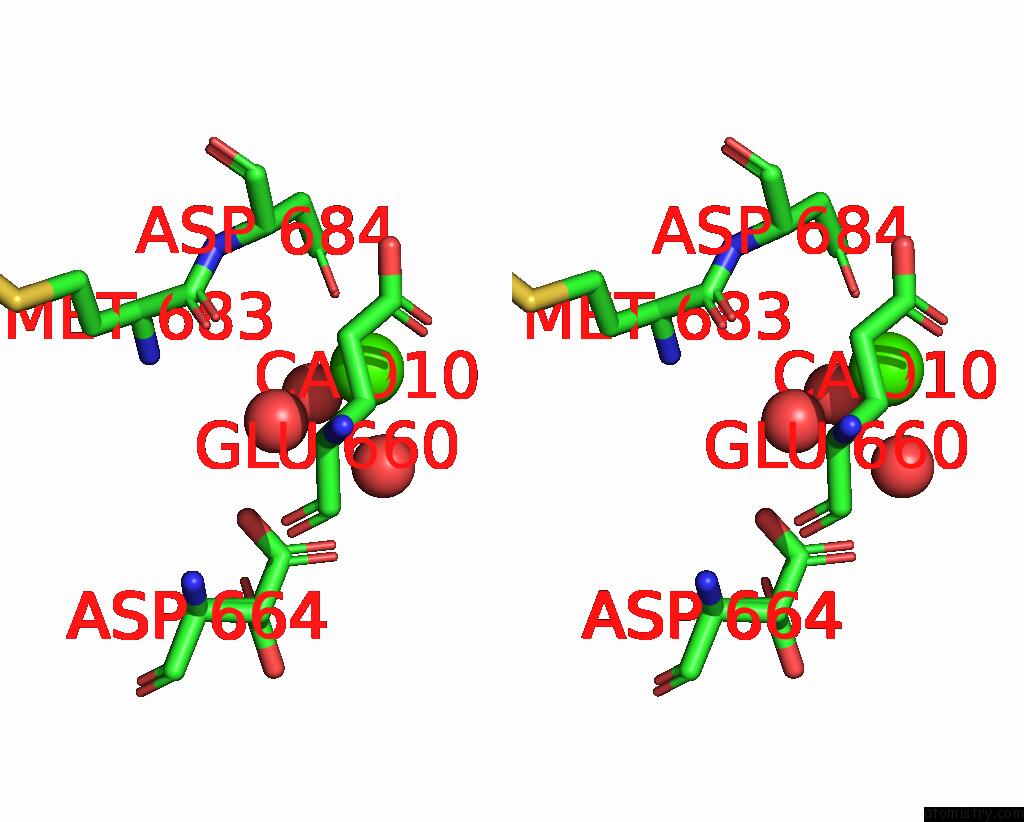
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 6 of RB69 Dna Polymerase (S565G/Y567A) Ternary Complex with Dttp Opposite Dg within 5.0Å range:
|
Reference:
S.Xia,
M.Wang,
H.R.Lee,
A.Sinha,
G.Blaha,
T.Christian,
J.Wang,
W.Konigsberg.
Variation in Mutation Rates Caused By RB69POL Fidelity Mutants Can Be Rationalized on the Basis of Their Kinetic Behavior and Crystal Structures. J.Mol.Biol. V. 406 558 2011.
ISSN: ISSN 0022-2836
PubMed: 21216248
DOI: 10.1016/J.JMB.2010.12.033
Page generated: Sat Jul 13 15:00:00 2024
ISSN: ISSN 0022-2836
PubMed: 21216248
DOI: 10.1016/J.JMB.2010.12.033
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1