Calcium »
PDB 3p1o-3pdd »
3p5c »
Calcium in PDB 3p5c: The Structure of the Ldlr/PCSK9 Complex Reveals the Receptor in An Extended Conformation
Protein crystallography data
The structure of The Structure of the Ldlr/PCSK9 Complex Reveals the Receptor in An Extended Conformation, PDB code: 3p5c
was solved by
P.Lo Surdo,
M.J.Bottomley,
A.Calzetta,
E.C.Settembre,
A.Cirillo,
S.Pandit,
Y.Ni,
B.Hubbard,
A.Sitlani,
A.Carfi,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 4.20 |
| Space group | P 65 |
| Cell size a, b, c (Å), α, β, γ (°) | 320.886, 320.886, 77.118, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 32.5 / 34.8 |
Calcium Binding Sites:
The binding sites of Calcium atom in the The Structure of the Ldlr/PCSK9 Complex Reveals the Receptor in An Extended Conformation
(pdb code 3p5c). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 4 binding sites of Calcium where determined in the The Structure of the Ldlr/PCSK9 Complex Reveals the Receptor in An Extended Conformation, PDB code: 3p5c:
Jump to Calcium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Calcium where determined in the The Structure of the Ldlr/PCSK9 Complex Reveals the Receptor in An Extended Conformation, PDB code: 3p5c:
Jump to Calcium binding site number: 1; 2; 3; 4;
Calcium binding site 1 out of 4 in 3p5c
Go back to
Calcium binding site 1 out
of 4 in the The Structure of the Ldlr/PCSK9 Complex Reveals the Receptor in An Extended Conformation
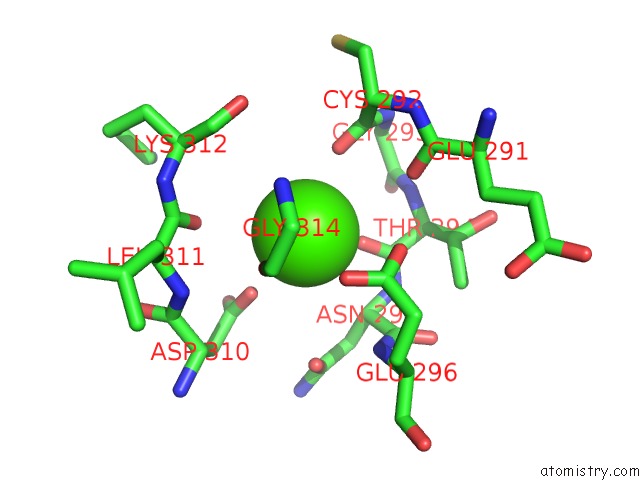
Mono view
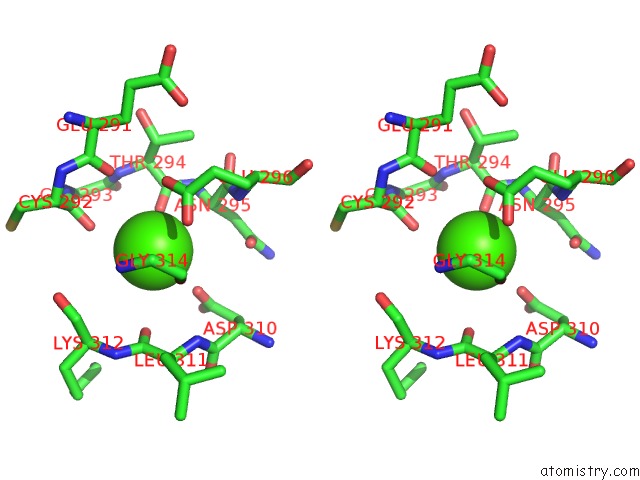
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of The Structure of the Ldlr/PCSK9 Complex Reveals the Receptor in An Extended Conformation within 5.0Å range:
|
Calcium binding site 2 out of 4 in 3p5c
Go back to
Calcium binding site 2 out
of 4 in the The Structure of the Ldlr/PCSK9 Complex Reveals the Receptor in An Extended Conformation
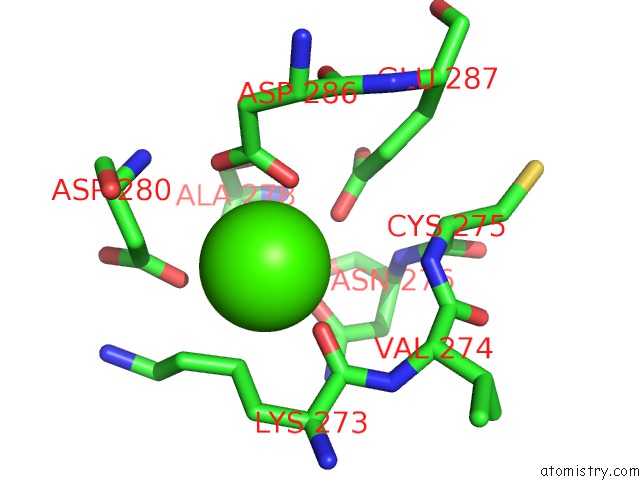
Mono view
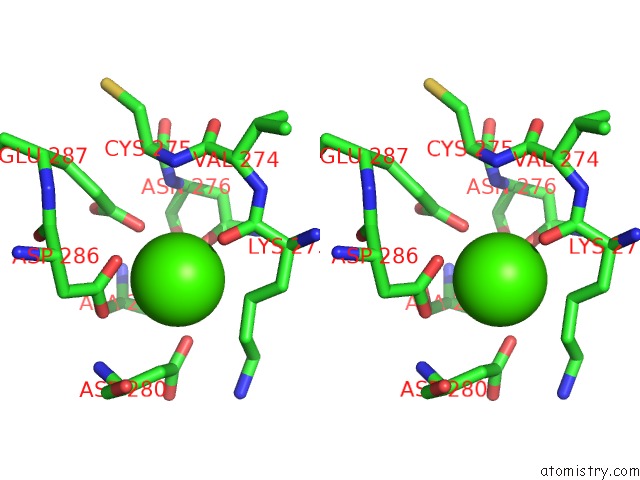
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of The Structure of the Ldlr/PCSK9 Complex Reveals the Receptor in An Extended Conformation within 5.0Å range:
|
Calcium binding site 3 out of 4 in 3p5c
Go back to
Calcium binding site 3 out
of 4 in the The Structure of the Ldlr/PCSK9 Complex Reveals the Receptor in An Extended Conformation
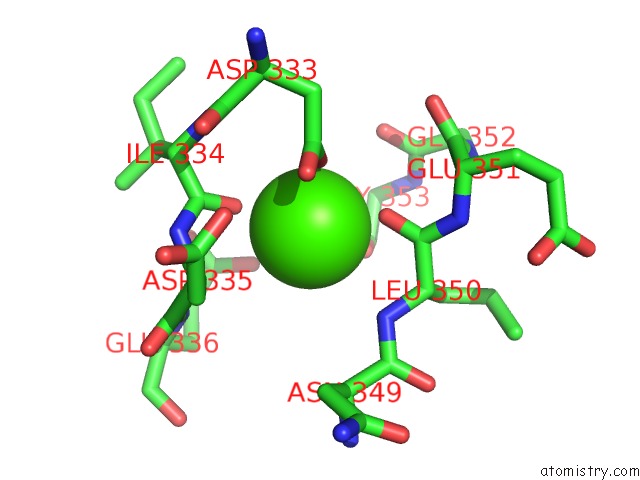
Mono view
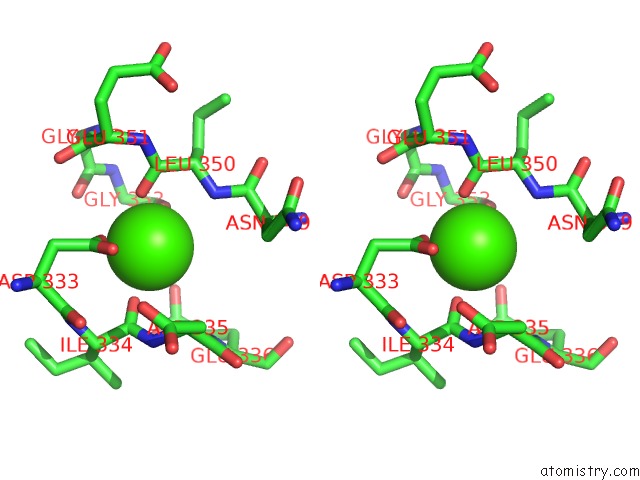
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of The Structure of the Ldlr/PCSK9 Complex Reveals the Receptor in An Extended Conformation within 5.0Å range:
|
Calcium binding site 4 out of 4 in 3p5c
Go back to
Calcium binding site 4 out
of 4 in the The Structure of the Ldlr/PCSK9 Complex Reveals the Receptor in An Extended Conformation
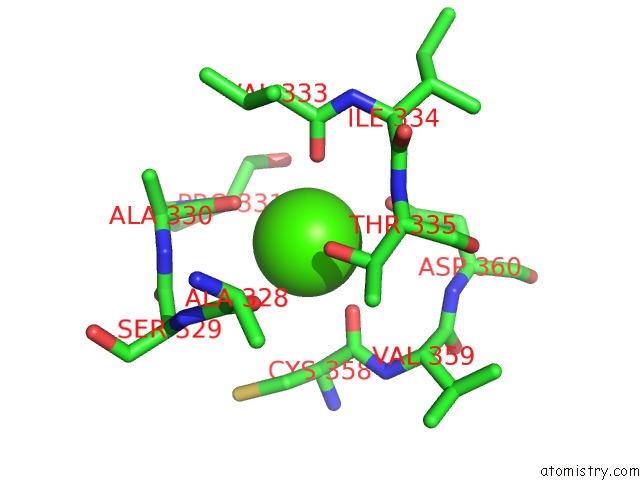
Mono view
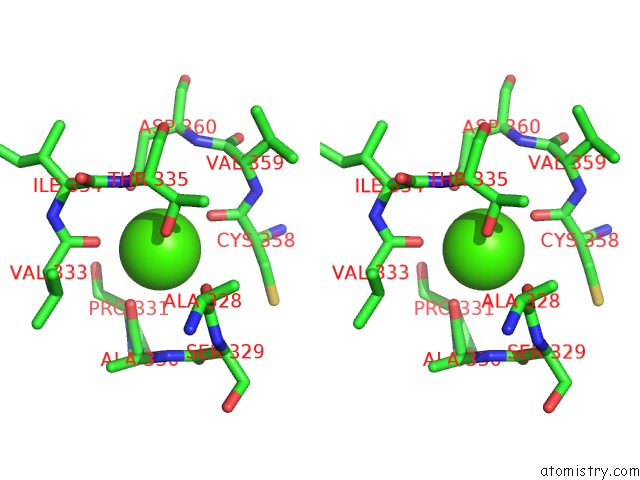
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of The Structure of the Ldlr/PCSK9 Complex Reveals the Receptor in An Extended Conformation within 5.0Å range:
|
Reference:
P.Lo Surdo,
M.J.Bottomley,
A.Calzetta,
E.C.Settembre,
A.Cirillo,
S.Pandit,
Y.G.Ni,
B.Hubbard,
A.Sitlani,
A.Carfi.
Mechanistic Implications For Ldl Receptor Degradation From the PCSK9/Ldlr Structure at Neutral pH. Embo Rep. V. 12 1300 2011.
ISSN: ISSN 1469-221X
PubMed: 22081141
DOI: 10.1038/EMBOR.2011.205
Page generated: Tue Jul 8 15:16:38 2025
ISSN: ISSN 1469-221X
PubMed: 22081141
DOI: 10.1038/EMBOR.2011.205
Last articles
F in 4EPXF in 4ENC
F in 4ENB
F in 4EMV
F in 4ENA
F in 4EN5
F in 4EKC
F in 4EKD
F in 4EHG
F in 4EHE