Calcium »
PDB 3rxb-3s9n »
3s55 »
Calcium in PDB 3s55: Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad
Protein crystallography data
The structure of Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad, PDB code: 3s55
was solved by
Seattle Structural Genomics Center For Infectious Disease (Ssgcid),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.10 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 69.360, 84.970, 100.890, 81.77, 76.78, 74.23 |
| R / Rfree (%) | 18.9 / 23.7 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad
(pdb code 3s55). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 8 binding sites of Calcium where determined in the Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad, PDB code: 3s55:
Jump to Calcium binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Calcium where determined in the Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad, PDB code: 3s55:
Jump to Calcium binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Calcium binding site 1 out of 8 in 3s55
Go back to
Calcium binding site 1 out
of 8 in the Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad
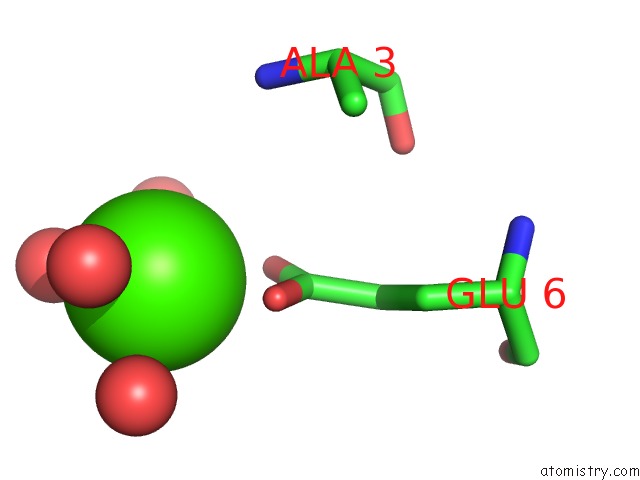
Mono view
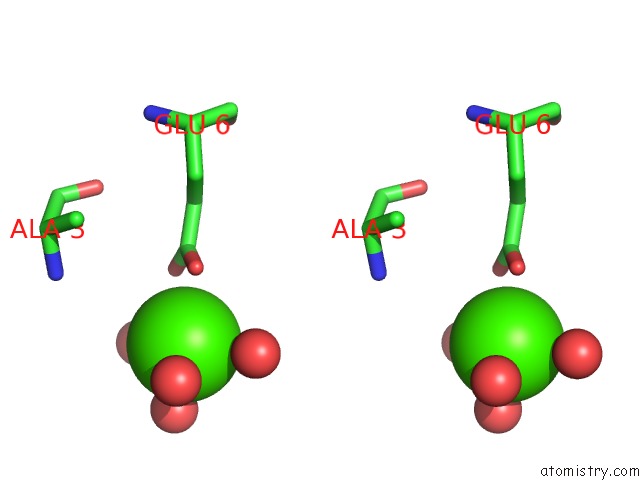
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad within 5.0Å range:
|
Calcium binding site 2 out of 8 in 3s55
Go back to
Calcium binding site 2 out
of 8 in the Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad
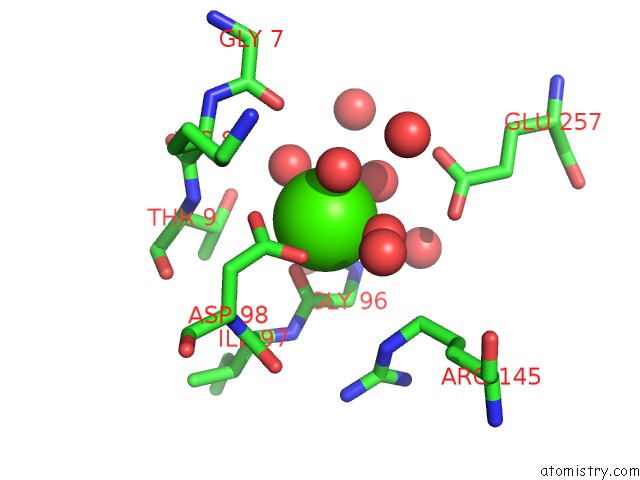
Mono view
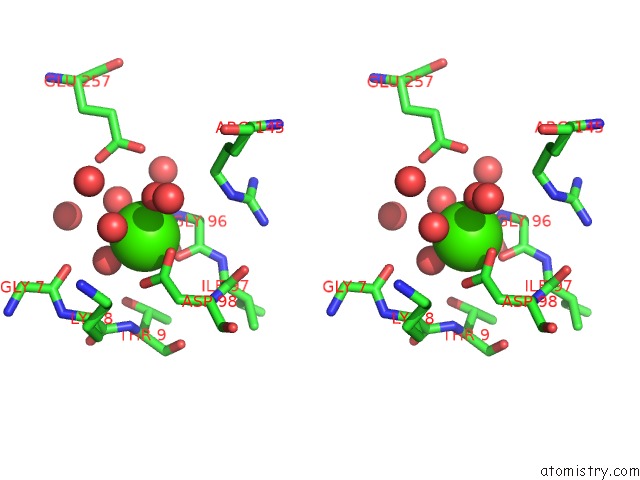
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad within 5.0Å range:
|
Calcium binding site 3 out of 8 in 3s55
Go back to
Calcium binding site 3 out
of 8 in the Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad
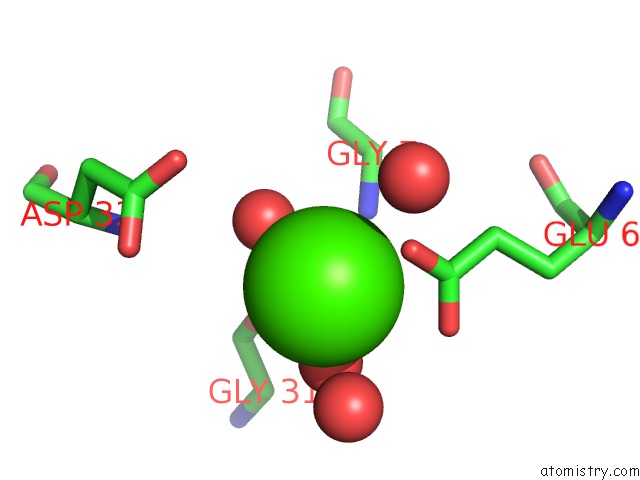
Mono view
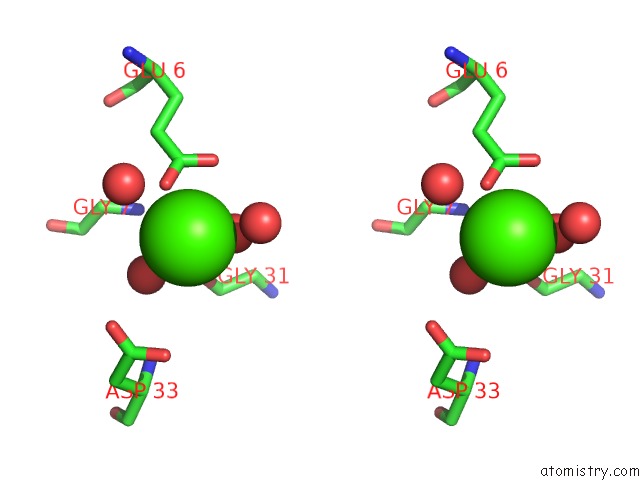
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad within 5.0Å range:
|
Calcium binding site 4 out of 8 in 3s55
Go back to
Calcium binding site 4 out
of 8 in the Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad
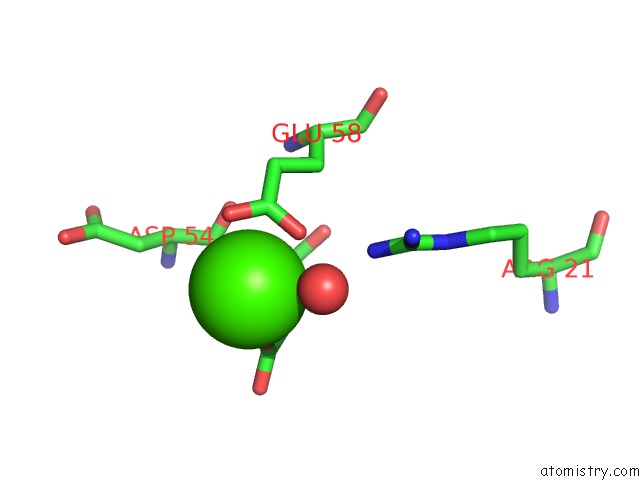
Mono view
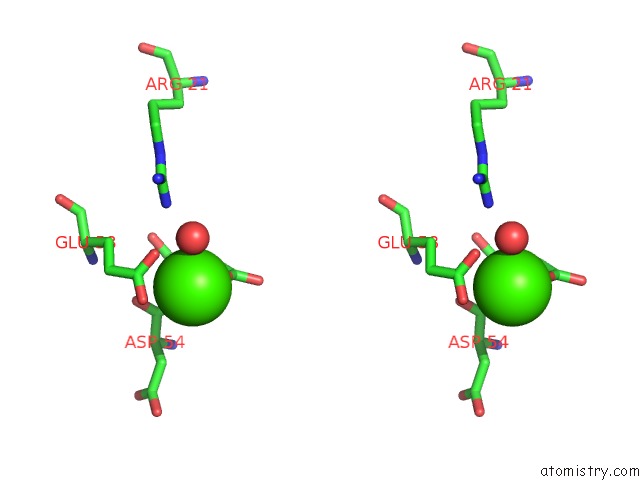
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad within 5.0Å range:
|
Calcium binding site 5 out of 8 in 3s55
Go back to
Calcium binding site 5 out
of 8 in the Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad
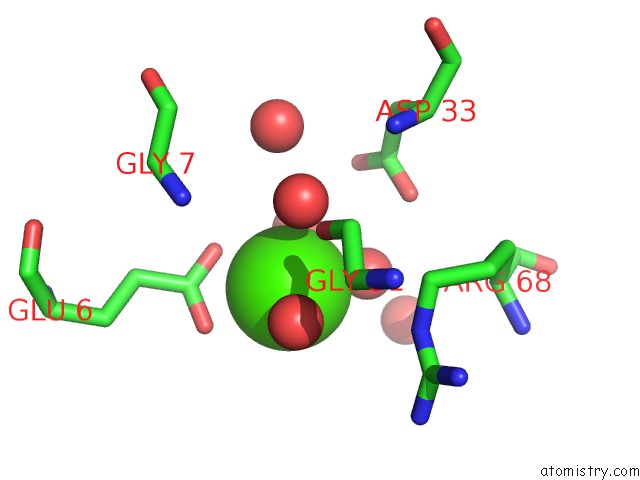
Mono view
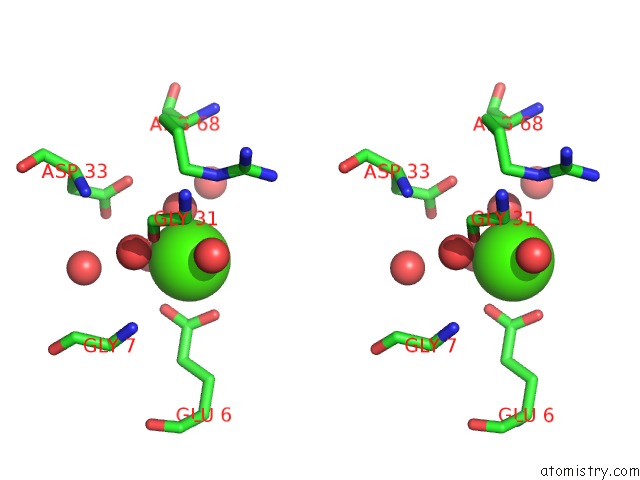
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 5 of Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad within 5.0Å range:
|
Calcium binding site 6 out of 8 in 3s55
Go back to
Calcium binding site 6 out
of 8 in the Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad
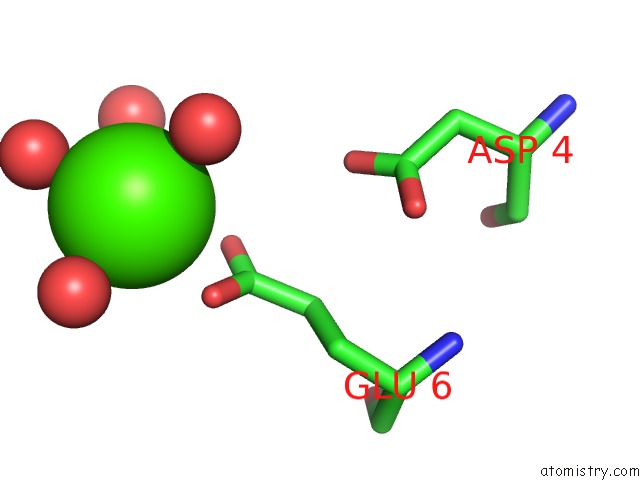
Mono view
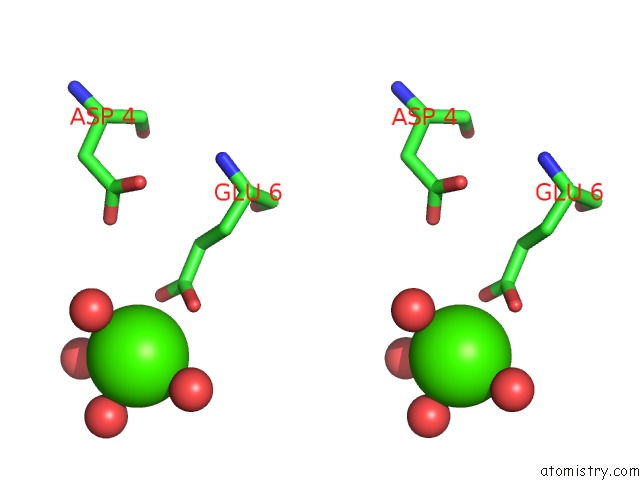
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 6 of Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad within 5.0Å range:
|
Calcium binding site 7 out of 8 in 3s55
Go back to
Calcium binding site 7 out
of 8 in the Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad
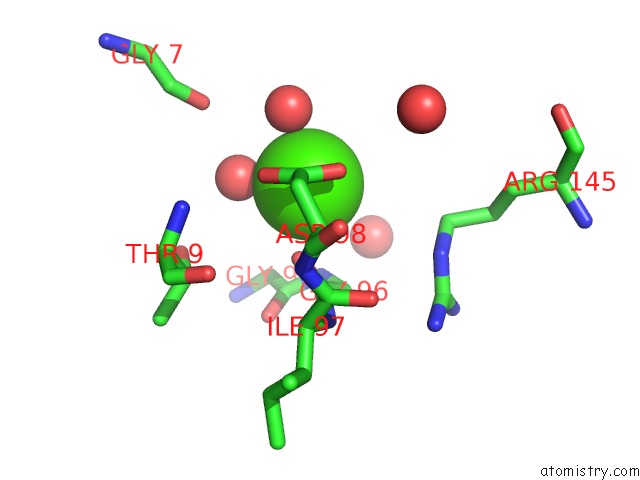
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 7 of Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad within 5.0Å range:
|
Calcium binding site 8 out of 8 in 3s55
Go back to
Calcium binding site 8 out
of 8 in the Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad
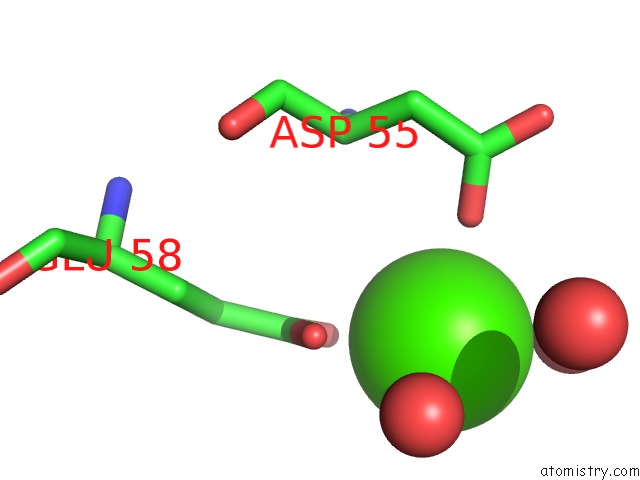
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 8 of Crystal Structure of A Putative Short-Chain Dehydrogenase/Reductase From Mycobacterium Abscessus Bound to Nad within 5.0Å range:
|
Reference:
D.H.Haft,
P.G.Pierce,
S.J.Mayclin,
A.Sullivan,
A.S.Gardberg,
J.Abendroth,
D.W.Begley,
I.Q.Phan,
B.L.Staker,
P.J.Myler,
V.M.Marathias,
D.D.Lorimer,
T.E.Edwards.
Mycofactocin-Associated Mycobacterial Dehydrogenases with Non-Exchangeable Nad Cofactors. Sci Rep V. 7 41074 2017.
ISSN: ESSN 2045-2322
PubMed: 28120876
DOI: 10.1038/SREP41074
Page generated: Tue Jul 8 16:29:55 2025
ISSN: ESSN 2045-2322
PubMed: 28120876
DOI: 10.1038/SREP41074
Last articles
F in 7MXNF in 7MXG
F in 7MXH
F in 7MX7
F in 7MVS
F in 7MX0
F in 7MXA
F in 7MT6
F in 7MSA
F in 7MTY