Calcium »
PDB 3so0-3t2i »
3sxq »
Calcium in PDB 3sxq: Structure of A Hexameric Multiheme C Nitrite Reductase From the Extremophile Bacterium Thiolkalivibrio Paradoxus
Protein crystallography data
The structure of Structure of A Hexameric Multiheme C Nitrite Reductase From the Extremophile Bacterium Thiolkalivibrio Paradoxus, PDB code: 3sxq
was solved by
K.M.Polyakov,
A.A.Trofimov,
T.V.Tikhonova,
A.V.Tikhonov,
K.M.Boyko,
V.O.Popov,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.90 / 1.90 |
| Space group | P 21 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 193.640, 193.640, 193.640, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 13.8 / 16.2 |
Other elements in 3sxq:
The structure of Structure of A Hexameric Multiheme C Nitrite Reductase From the Extremophile Bacterium Thiolkalivibrio Paradoxus also contains other interesting chemical elements:
| Cobalt | (Co) | 4 atoms |
| Iron | (Fe) | 16 atoms |
| Chlorine | (Cl) | 2 atoms |
| Sodium | (Na) | 2 atoms |
Calcium Binding Sites:
The binding sites of Calcium atom in the Structure of A Hexameric Multiheme C Nitrite Reductase From the Extremophile Bacterium Thiolkalivibrio Paradoxus
(pdb code 3sxq). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Structure of A Hexameric Multiheme C Nitrite Reductase From the Extremophile Bacterium Thiolkalivibrio Paradoxus, PDB code: 3sxq:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Structure of A Hexameric Multiheme C Nitrite Reductase From the Extremophile Bacterium Thiolkalivibrio Paradoxus, PDB code: 3sxq:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 3sxq
Go back to
Calcium binding site 1 out
of 2 in the Structure of A Hexameric Multiheme C Nitrite Reductase From the Extremophile Bacterium Thiolkalivibrio Paradoxus

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Structure of A Hexameric Multiheme C Nitrite Reductase From the Extremophile Bacterium Thiolkalivibrio Paradoxus within 5.0Å range:
|
Calcium binding site 2 out of 2 in 3sxq
Go back to
Calcium binding site 2 out
of 2 in the Structure of A Hexameric Multiheme C Nitrite Reductase From the Extremophile Bacterium Thiolkalivibrio Paradoxus
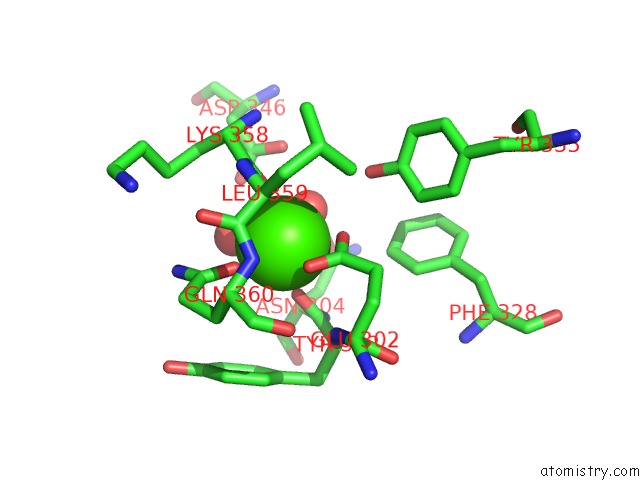
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Structure of A Hexameric Multiheme C Nitrite Reductase From the Extremophile Bacterium Thiolkalivibrio Paradoxus within 5.0Å range:
|
Reference:
T.Tikhonova,
A.Tikhonov,
A.Trofimov,
K.Polyakov,
K.Boyko,
E.Cherkashin,
T.Rakitina,
D.Sorokin,
V.Popov.
Comparative Structural and Functional Analysis of Two Octaheme Nitrite Reductases From Closely Related Thioalkalivibrio Species. Febs J. V. 279 4052 2012.
ISSN: ISSN 1742-464X
PubMed: 22935005
DOI: 10.1111/J.1742-4658.2012.08811.X
Page generated: Tue Jul 8 16:47:38 2025
ISSN: ISSN 1742-464X
PubMed: 22935005
DOI: 10.1111/J.1742-4658.2012.08811.X
Last articles
Ca in 7OR8Ca in 7OFQ
Ca in 7OR1
Ca in 7ONG
Ca in 7OR0
Ca in 7OP2
Ca in 7OO7
Ca in 7OO5
Ca in 7O85
Ca in 7OIH