Calcium »
PDB 3ti4-3tvc »
3ts5 »
Calcium in PDB 3ts5: Crystal Structure of A Light Chain Domain of Scallop Smooth Muscle Myosin
Protein crystallography data
The structure of Crystal Structure of A Light Chain Domain of Scallop Smooth Muscle Myosin, PDB code: 3ts5
was solved by
V.S.S.Kumar,
E.O'neall-Hennessey,
L.Reshetnikova,
J.H.Brown,
H.Robinson,
A.G.Szent-Gyorgyi,
C.Cohen,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.75 / 2.39 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 50.811, 68.775, 79.418, 77.33, 85.99, 73.65 |
| R / Rfree (%) | 19.1 / 24.1 |
Other elements in 3ts5:
The structure of Crystal Structure of A Light Chain Domain of Scallop Smooth Muscle Myosin also contains other interesting chemical elements:
| Magnesium | (Mg) | 2 atoms |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of A Light Chain Domain of Scallop Smooth Muscle Myosin
(pdb code 3ts5). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Crystal Structure of A Light Chain Domain of Scallop Smooth Muscle Myosin, PDB code: 3ts5:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Crystal Structure of A Light Chain Domain of Scallop Smooth Muscle Myosin, PDB code: 3ts5:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 3ts5
Go back to
Calcium binding site 1 out
of 2 in the Crystal Structure of A Light Chain Domain of Scallop Smooth Muscle Myosin
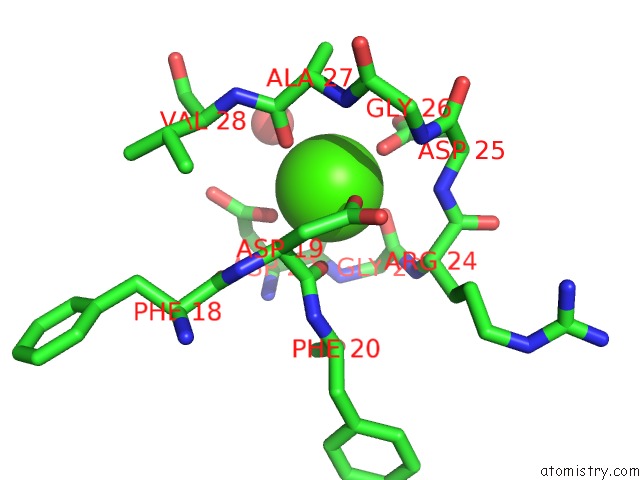
Mono view
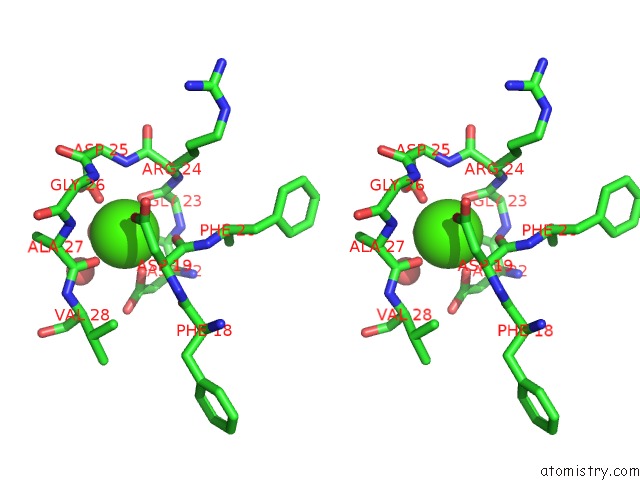
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of A Light Chain Domain of Scallop Smooth Muscle Myosin within 5.0Å range:
|
Calcium binding site 2 out of 2 in 3ts5
Go back to
Calcium binding site 2 out
of 2 in the Crystal Structure of A Light Chain Domain of Scallop Smooth Muscle Myosin

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Crystal Structure of A Light Chain Domain of Scallop Smooth Muscle Myosin within 5.0Å range:
|
Reference:
V.S.Senthil Kumar,
E.O'neall-Hennessey,
L.Reshetnikova,
J.H.Brown,
H.Robinson,
A.G.Szent-Gyorgyi,
C.Cohen.
Crystal Structure of A Phosphorylated Light Chain Domain of Scallop Smooth-Muscle Myosin. Biophys.J. V. 101 2185 2011.
ISSN: ISSN 0006-3495
PubMed: 22067157
DOI: 10.1016/J.BPJ.2011.09.028
Page generated: Tue Jul 8 17:04:32 2025
ISSN: ISSN 0006-3495
PubMed: 22067157
DOI: 10.1016/J.BPJ.2011.09.028
Last articles
F in 4IJUF in 4IKL
F in 4IJV
F in 4IKK
F in 4IKJ
F in 4IK6
F in 4IKF
F in 4IJH
F in 4IGS
F in 4IF4