Calcium »
PDB 3uix-3v03 »
3usn »
Calcium in PDB 3usn: Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure
Enzymatic activity of Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure
All present enzymatic activity of Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure:
3.4.24.17;
3.4.24.17;
Other elements in 3usn:
The structure of Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
Calcium Binding Sites:
The binding sites of Calcium atom in the Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure
(pdb code 3usn). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 3 binding sites of Calcium where determined in the Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure, PDB code: 3usn:
Jump to Calcium binding site number: 1; 2; 3;
In total 3 binding sites of Calcium where determined in the Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure, PDB code: 3usn:
Jump to Calcium binding site number: 1; 2; 3;
Calcium binding site 1 out of 3 in 3usn
Go back to
Calcium binding site 1 out
of 3 in the Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure
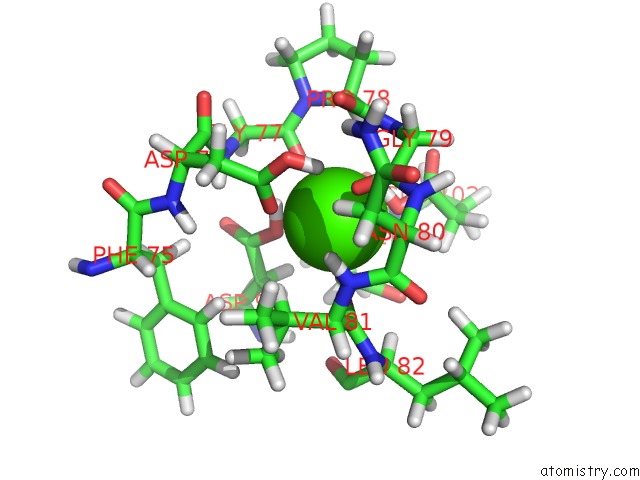
Mono view
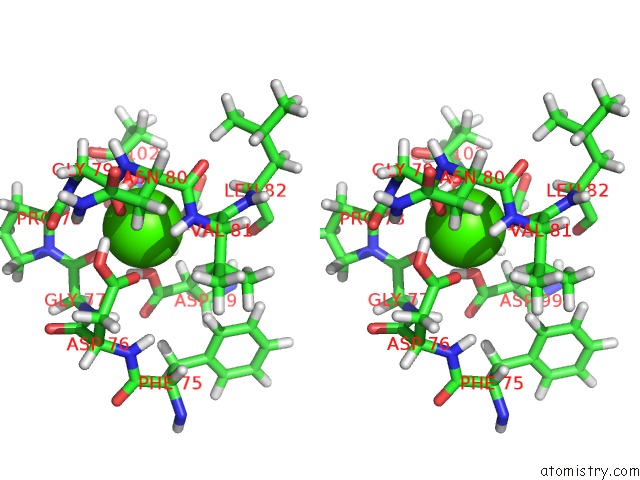
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure within 5.0Å range:
|
Calcium binding site 2 out of 3 in 3usn
Go back to
Calcium binding site 2 out
of 3 in the Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure
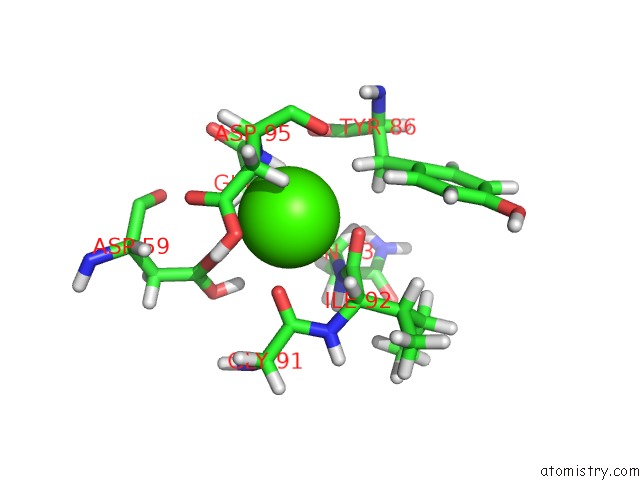
Mono view
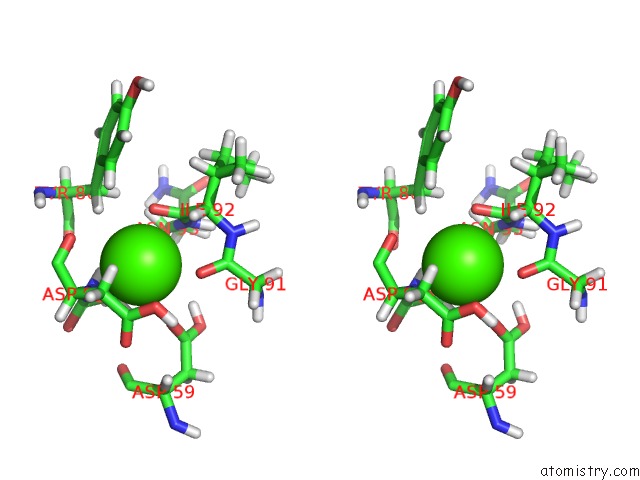
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure within 5.0Å range:
|
Calcium binding site 3 out of 3 in 3usn
Go back to
Calcium binding site 3 out
of 3 in the Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure
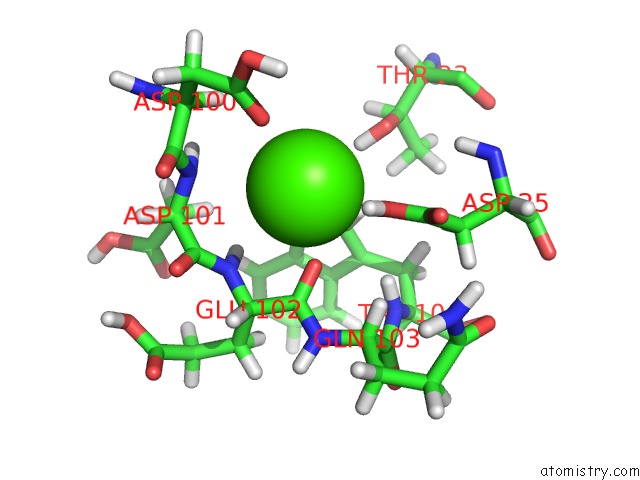
Mono view
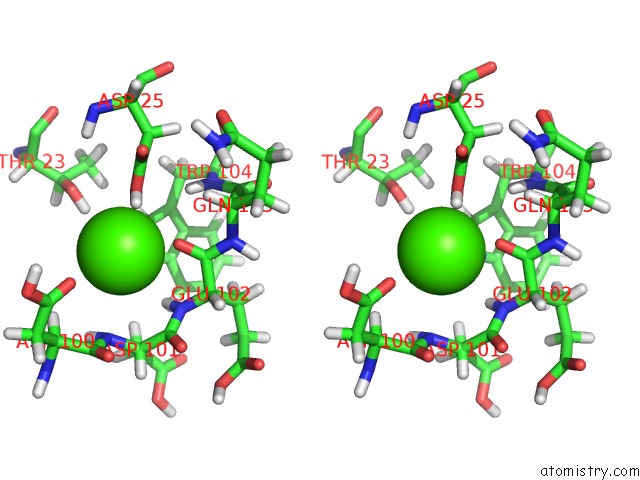
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure within 5.0Å range:
|
Reference:
B.J.Stockman,
D.J.Waldon,
J.A.Gates,
T.A.Scahill,
D.A.Kloosterman,
S.A.Mizsak,
E.J.Jacobsen,
K.L.Belonga,
M.A.Mitchell,
B.Mao,
J.D.Petke,
L.Goodman,
E.A.Powers,
S.R.Ledbetter,
P.S.Kaytes,
G.Vogeli,
V.P.Marshall,
G.L.Petzold,
R.A.Poorman.
Solution Structures of Stromelysin Complexed to Thiadiazole Inhibitors. Protein Sci. V. 7 2281 1998.
ISSN: ISSN 0961-8368
PubMed: 9827994
Page generated: Tue Jul 8 17:19:51 2025
ISSN: ISSN 0961-8368
PubMed: 9827994
Last articles
F in 4G2IF in 4G2H
F in 4G31
F in 4G1W
F in 4G16
F in 4FXY
F in 4FXQ
F in 4FZF
F in 4FP1
F in 4FVX