Calcium »
PDB 4eoy-4fb1 »
4esv »
Calcium in PDB 4esv: A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates
Enzymatic activity of A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates
All present enzymatic activity of A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates:
3.6.4.12;
3.6.4.12;
Protein crystallography data
The structure of A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates, PDB code: 4esv
was solved by
O.Itsathitphaisarn,
R.A.Wing,
W.K.Eliason,
J.Wang,
T.A.Steitz,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.92 / 3.20 |
| Space group | P 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 149.124, 180.321, 279.128, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 24.2 / 28.9 |
Other elements in 4esv:
The structure of A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates also contains other interesting chemical elements:
| Fluorine | (F) | 40 atoms |
| Aluminium | (Al) | 10 atoms |
Calcium Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 14;Binding sites:
The binding sites of Calcium atom in the A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates (pdb code 4esv). This binding sites where shown within 5.0 Angstroms radius around Calcium atom.In total 14 binding sites of Calcium where determined in the A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates, PDB code: 4esv:
Jump to Calcium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Calcium binding site 1 out of 14 in 4esv
Go back to
Calcium binding site 1 out
of 14 in the A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates
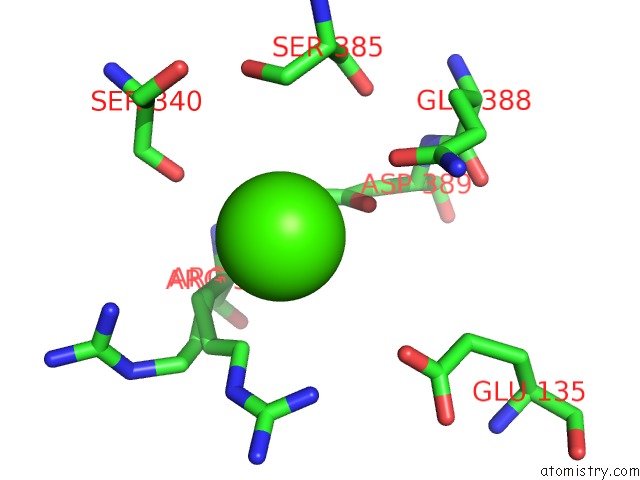
Mono view
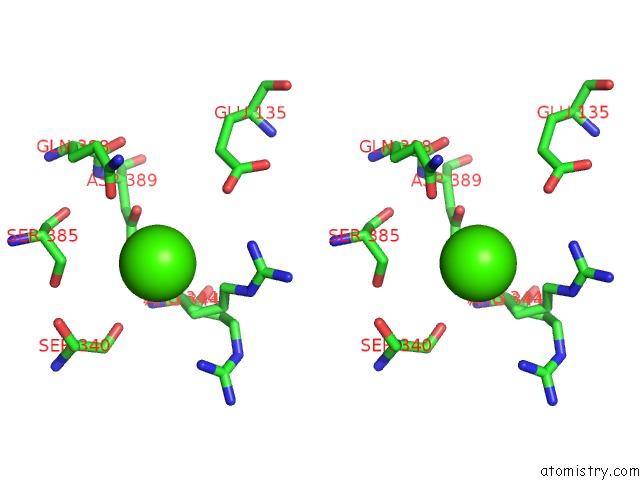
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates within 5.0Å range:
|
Calcium binding site 2 out of 14 in 4esv
Go back to
Calcium binding site 2 out
of 14 in the A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates within 5.0Å range:
|
Calcium binding site 3 out of 14 in 4esv
Go back to
Calcium binding site 3 out
of 14 in the A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates

Mono view
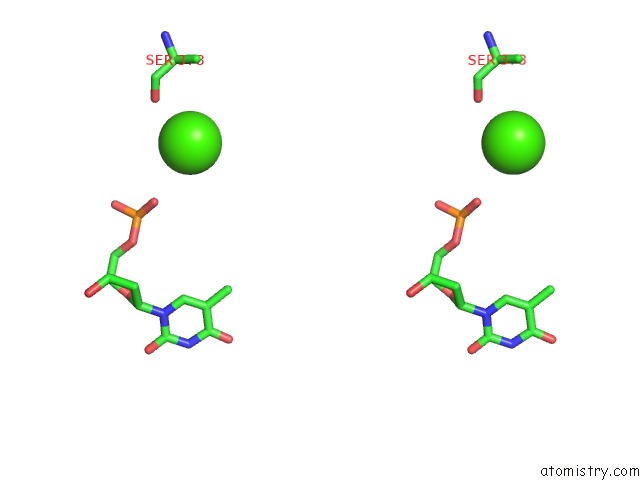
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates within 5.0Å range:
|
Calcium binding site 4 out of 14 in 4esv
Go back to
Calcium binding site 4 out
of 14 in the A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates
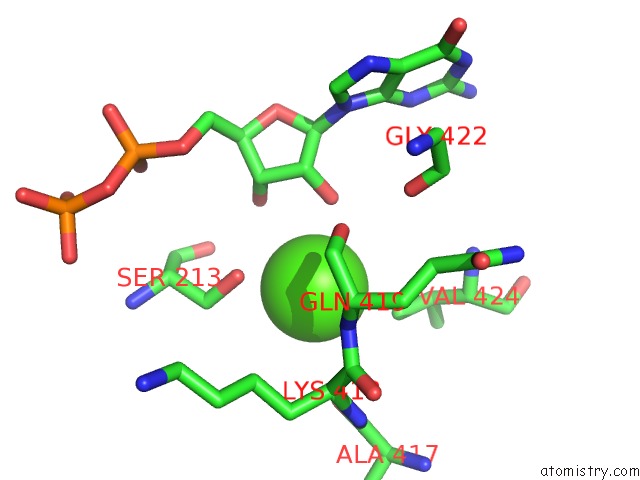
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates within 5.0Å range:
|
Calcium binding site 5 out of 14 in 4esv
Go back to
Calcium binding site 5 out
of 14 in the A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 5 of A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates within 5.0Å range:
|
Calcium binding site 6 out of 14 in 4esv
Go back to
Calcium binding site 6 out
of 14 in the A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates

Mono view
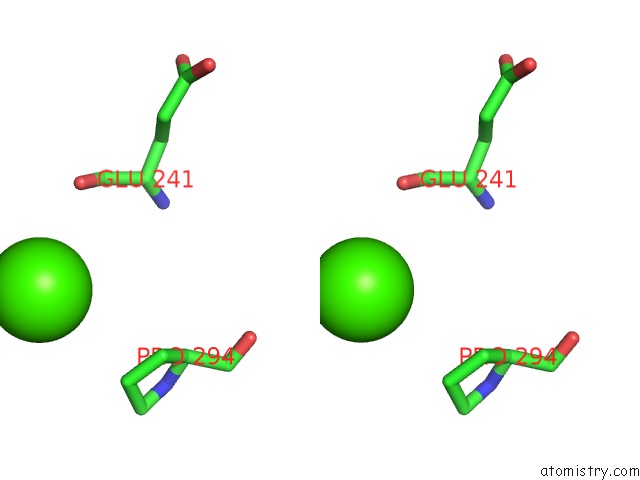
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 6 of A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates within 5.0Å range:
|
Calcium binding site 7 out of 14 in 4esv
Go back to
Calcium binding site 7 out
of 14 in the A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates
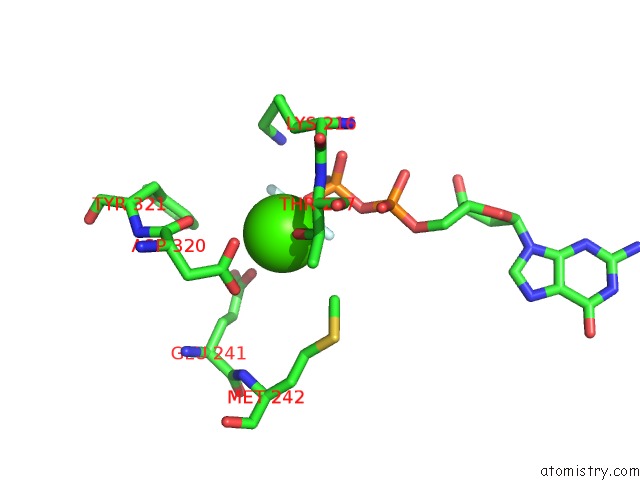
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 7 of A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates within 5.0Å range:
|
Calcium binding site 8 out of 14 in 4esv
Go back to
Calcium binding site 8 out
of 14 in the A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 8 of A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates within 5.0Å range:
|
Calcium binding site 9 out of 14 in 4esv
Go back to
Calcium binding site 9 out
of 14 in the A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates
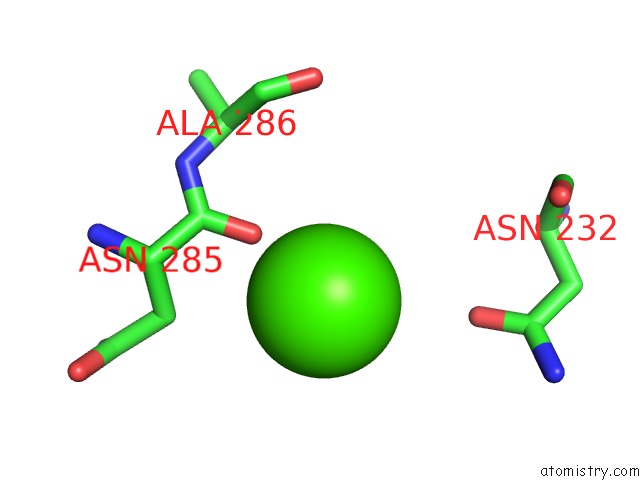
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 9 of A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates within 5.0Å range:
|
Calcium binding site 10 out of 14 in 4esv
Go back to
Calcium binding site 10 out
of 14 in the A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates

Mono view
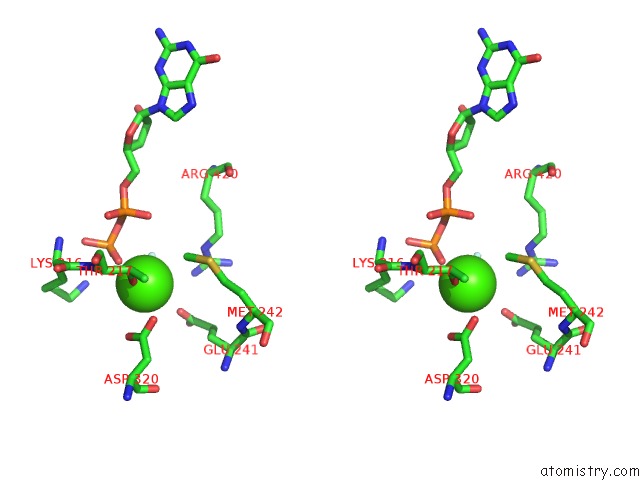
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 10 of A New Twist on the Translocation Mechanism of Helicases From the Structure of Dnab with Its Substrates within 5.0Å range:
|
Reference:
O.Itsathitphaisarn,
R.A.Wing,
W.K.Eliason,
J.Wang,
T.A.Steitz.
The Hexameric Helicase Dnab Adopts A Nonplanar Conformation During Translocation. Cell(Cambridge,Mass.) V. 151 267 2012.
ISSN: ISSN 0092-8674
PubMed: 23022319
DOI: 10.1016/J.CELL.2012.09.014
Page generated: Tue Jul 8 19:52:00 2025
ISSN: ISSN 0092-8674
PubMed: 23022319
DOI: 10.1016/J.CELL.2012.09.014
Last articles
Cl in 5HM0Cl in 5HLY
Cl in 5HLV
Cl in 5HLW
Cl in 5HLN
Cl in 5HLL
Cl in 5HLS
Cl in 5HKO
Cl in 5HKA
Cl in 5HLI