Calcium »
PDB 4ko2-4l19 »
4l04 »
Calcium in PDB 4l04: Crystal Structure Analysis of Human IDH1 Mutants in Complex with Nadp+ and CA2+/Alpha-Ketoglutarate
Enzymatic activity of Crystal Structure Analysis of Human IDH1 Mutants in Complex with Nadp+ and CA2+/Alpha-Ketoglutarate
All present enzymatic activity of Crystal Structure Analysis of Human IDH1 Mutants in Complex with Nadp+ and CA2+/Alpha-Ketoglutarate:
1.1.1.42;
1.1.1.42;
Protein crystallography data
The structure of Crystal Structure Analysis of Human IDH1 Mutants in Complex with Nadp+ and CA2+/Alpha-Ketoglutarate, PDB code: 4l04
was solved by
N.O.Concha,
A.M.Smallwood,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 49.24 / 2.87 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 96.410, 116.622, 275.729, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20 / 25.9 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure Analysis of Human IDH1 Mutants in Complex with Nadp+ and CA2+/Alpha-Ketoglutarate
(pdb code 4l04). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 6 binding sites of Calcium where determined in the Crystal Structure Analysis of Human IDH1 Mutants in Complex with Nadp+ and CA2+/Alpha-Ketoglutarate, PDB code: 4l04:
Jump to Calcium binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Calcium where determined in the Crystal Structure Analysis of Human IDH1 Mutants in Complex with Nadp+ and CA2+/Alpha-Ketoglutarate, PDB code: 4l04:
Jump to Calcium binding site number: 1; 2; 3; 4; 5; 6;
Calcium binding site 1 out of 6 in 4l04
Go back to
Calcium binding site 1 out
of 6 in the Crystal Structure Analysis of Human IDH1 Mutants in Complex with Nadp+ and CA2+/Alpha-Ketoglutarate
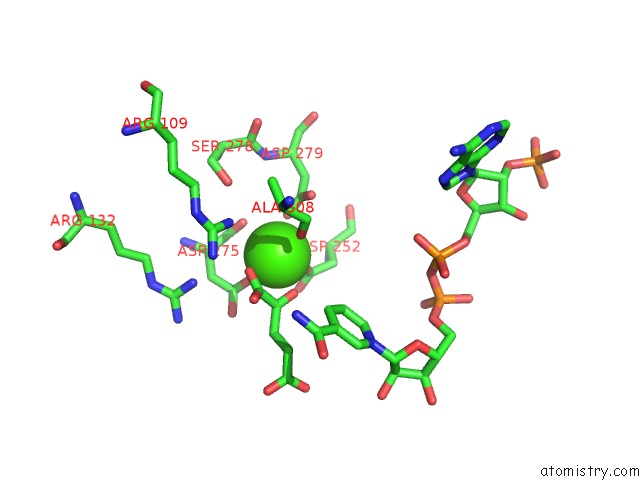
Mono view
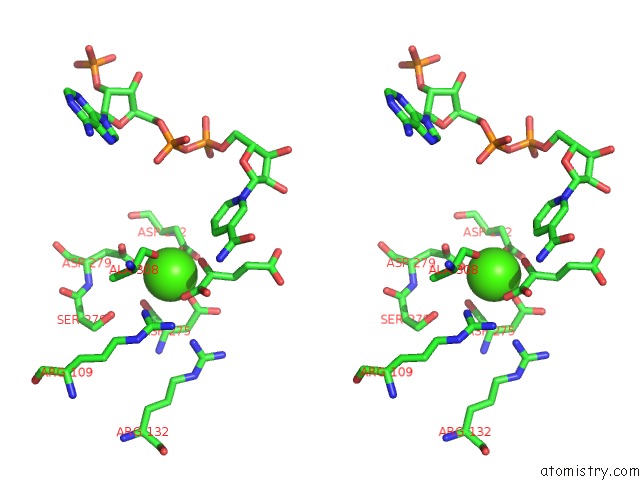
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure Analysis of Human IDH1 Mutants in Complex with Nadp+ and CA2+/Alpha-Ketoglutarate within 5.0Å range:
|
Calcium binding site 2 out of 6 in 4l04
Go back to
Calcium binding site 2 out
of 6 in the Crystal Structure Analysis of Human IDH1 Mutants in Complex with Nadp+ and CA2+/Alpha-Ketoglutarate
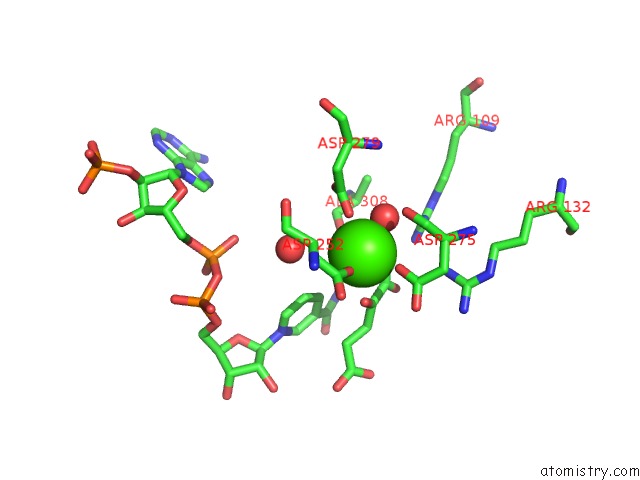
Mono view
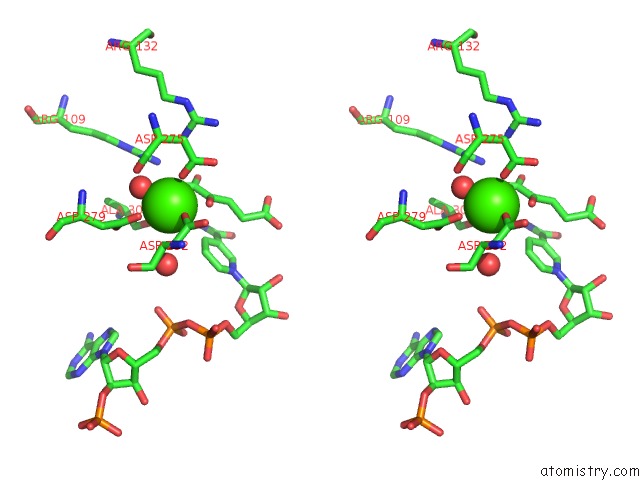
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Crystal Structure Analysis of Human IDH1 Mutants in Complex with Nadp+ and CA2+/Alpha-Ketoglutarate within 5.0Å range:
|
Calcium binding site 3 out of 6 in 4l04
Go back to
Calcium binding site 3 out
of 6 in the Crystal Structure Analysis of Human IDH1 Mutants in Complex with Nadp+ and CA2+/Alpha-Ketoglutarate
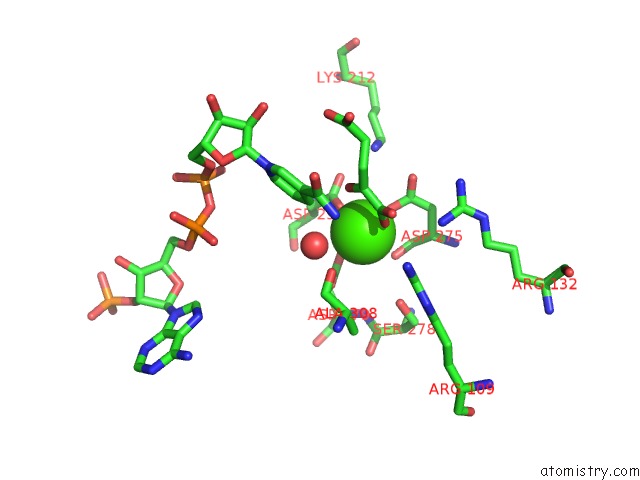
Mono view
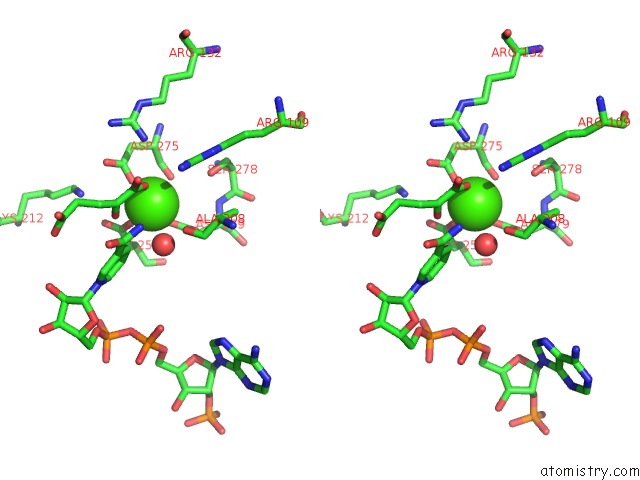
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Crystal Structure Analysis of Human IDH1 Mutants in Complex with Nadp+ and CA2+/Alpha-Ketoglutarate within 5.0Å range:
|
Calcium binding site 4 out of 6 in 4l04
Go back to
Calcium binding site 4 out
of 6 in the Crystal Structure Analysis of Human IDH1 Mutants in Complex with Nadp+ and CA2+/Alpha-Ketoglutarate
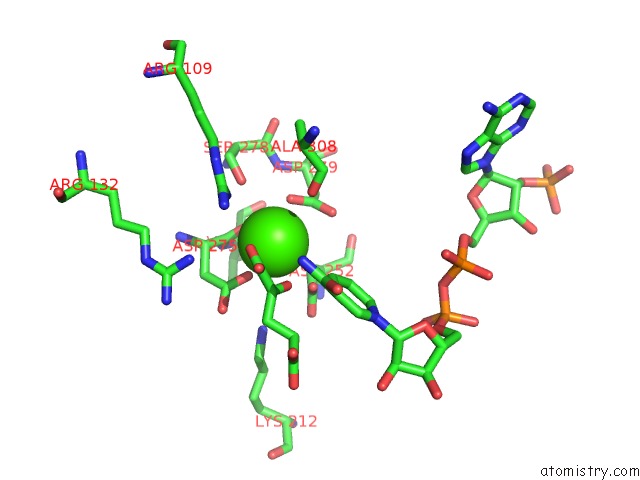
Mono view
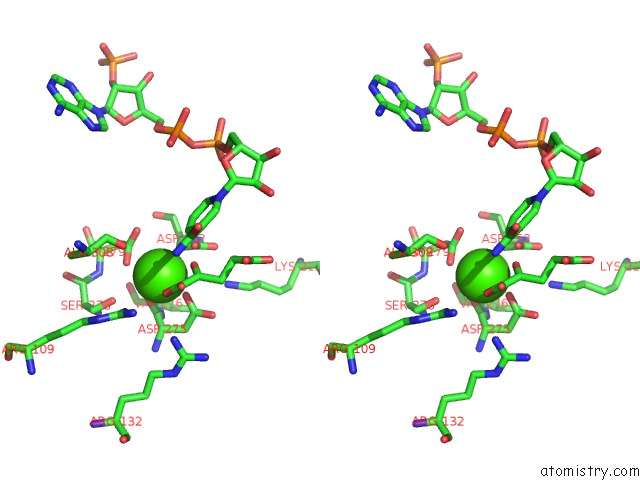
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of Crystal Structure Analysis of Human IDH1 Mutants in Complex with Nadp+ and CA2+/Alpha-Ketoglutarate within 5.0Å range:
|
Calcium binding site 5 out of 6 in 4l04
Go back to
Calcium binding site 5 out
of 6 in the Crystal Structure Analysis of Human IDH1 Mutants in Complex with Nadp+ and CA2+/Alpha-Ketoglutarate
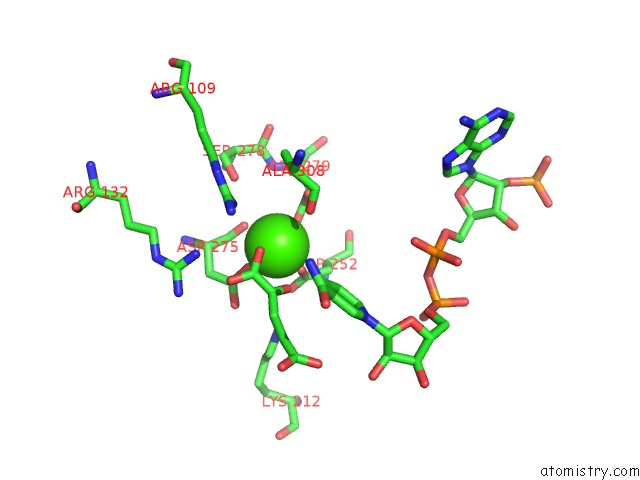
Mono view
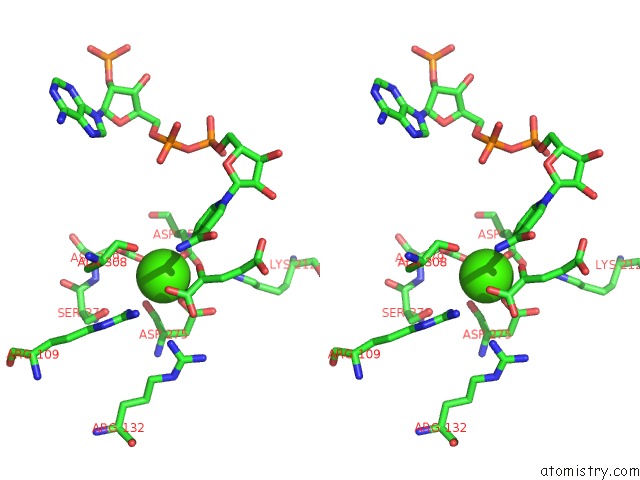
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 5 of Crystal Structure Analysis of Human IDH1 Mutants in Complex with Nadp+ and CA2+/Alpha-Ketoglutarate within 5.0Å range:
|
Calcium binding site 6 out of 6 in 4l04
Go back to
Calcium binding site 6 out
of 6 in the Crystal Structure Analysis of Human IDH1 Mutants in Complex with Nadp+ and CA2+/Alpha-Ketoglutarate
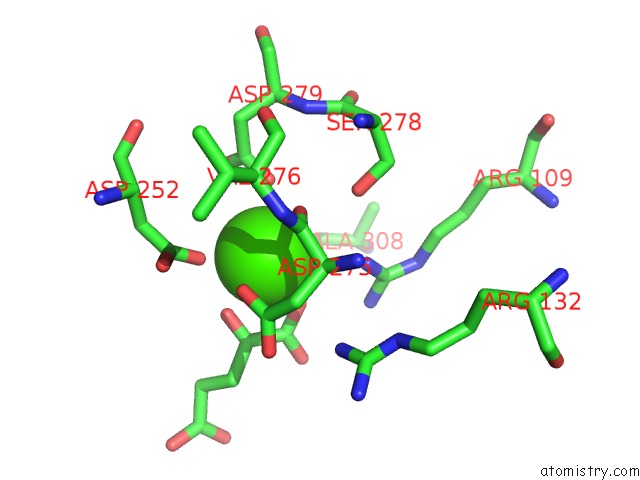
Mono view
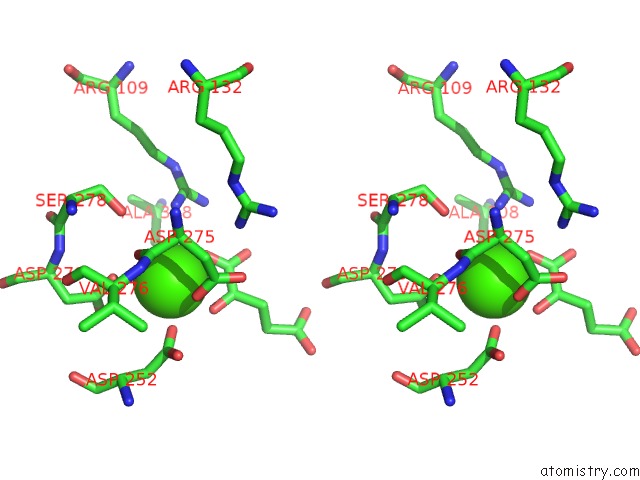
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 6 of Crystal Structure Analysis of Human IDH1 Mutants in Complex with Nadp+ and CA2+/Alpha-Ketoglutarate within 5.0Å range:
|
Reference:
A.R.Rendina,
B.Pietrak,
A.Smallwood,
H.Zhao,
H.Qi,
C.Quinn,
N.D.Adams,
N.Concha,
C.Duraiswami,
S.H.Thrall,
S.Sweitzer,
B.Schwartz.
Mutant IDH1 Enhances the Production of 2-Hydroxyglutarate Due to Its Kinetic Mechanism. Biochemistry V. 52 4563 2013.
ISSN: ISSN 0006-2960
PubMed: 23731180
DOI: 10.1021/BI400514K
Page generated: Tue Jul 8 23:43:27 2025
ISSN: ISSN 0006-2960
PubMed: 23731180
DOI: 10.1021/BI400514K
Last articles
Cl in 5G2CCl in 5G22
Cl in 5G27
Cl in 5G1Z
Cl in 5G0R
Cl in 5G0G
Cl in 5G1M
Cl in 5G0J
Cl in 5G0I
Cl in 5G08