Calcium »
PDB 4n0b-4n5x »
4n35 »
Calcium in PDB 4n35: Structure of Langerin Crd I313 Complexed with Glcnac-BETA1-3GAL-BETA1- 4GLC-Beta-CH2CH2N3
Protein crystallography data
The structure of Structure of Langerin Crd I313 Complexed with Glcnac-BETA1-3GAL-BETA1- 4GLC-Beta-CH2CH2N3, PDB code: 4n35
was solved by
H.Feinberg,
T.J.W.Rowntree,
S.L.W.Tan,
K.Drickamer,
W.I.Weis,
M.E.Taylor,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 35.58 / 1.85 |
| Space group | P 42 |
| Cell size a, b, c (Å), α, β, γ (°) | 79.490, 79.490, 91.930, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.6 / 21.2 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Structure of Langerin Crd I313 Complexed with Glcnac-BETA1-3GAL-BETA1- 4GLC-Beta-CH2CH2N3
(pdb code 4n35). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 4 binding sites of Calcium where determined in the Structure of Langerin Crd I313 Complexed with Glcnac-BETA1-3GAL-BETA1- 4GLC-Beta-CH2CH2N3, PDB code: 4n35:
Jump to Calcium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Calcium where determined in the Structure of Langerin Crd I313 Complexed with Glcnac-BETA1-3GAL-BETA1- 4GLC-Beta-CH2CH2N3, PDB code: 4n35:
Jump to Calcium binding site number: 1; 2; 3; 4;
Calcium binding site 1 out of 4 in 4n35
Go back to
Calcium binding site 1 out
of 4 in the Structure of Langerin Crd I313 Complexed with Glcnac-BETA1-3GAL-BETA1- 4GLC-Beta-CH2CH2N3
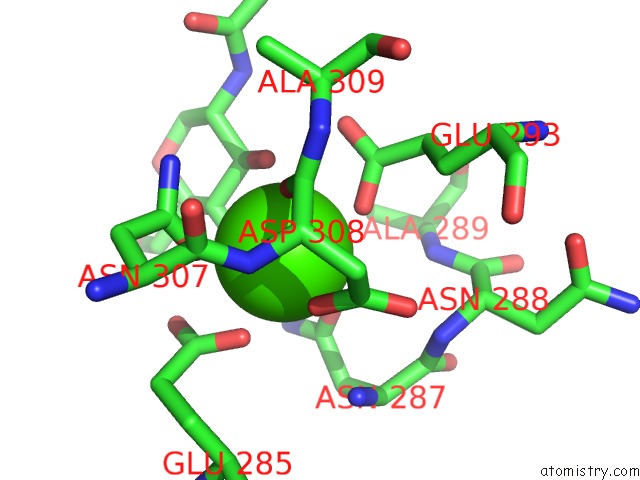
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Structure of Langerin Crd I313 Complexed with Glcnac-BETA1-3GAL-BETA1- 4GLC-Beta-CH2CH2N3 within 5.0Å range:
|
Calcium binding site 2 out of 4 in 4n35
Go back to
Calcium binding site 2 out
of 4 in the Structure of Langerin Crd I313 Complexed with Glcnac-BETA1-3GAL-BETA1- 4GLC-Beta-CH2CH2N3

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Structure of Langerin Crd I313 Complexed with Glcnac-BETA1-3GAL-BETA1- 4GLC-Beta-CH2CH2N3 within 5.0Å range:
|
Calcium binding site 3 out of 4 in 4n35
Go back to
Calcium binding site 3 out
of 4 in the Structure of Langerin Crd I313 Complexed with Glcnac-BETA1-3GAL-BETA1- 4GLC-Beta-CH2CH2N3
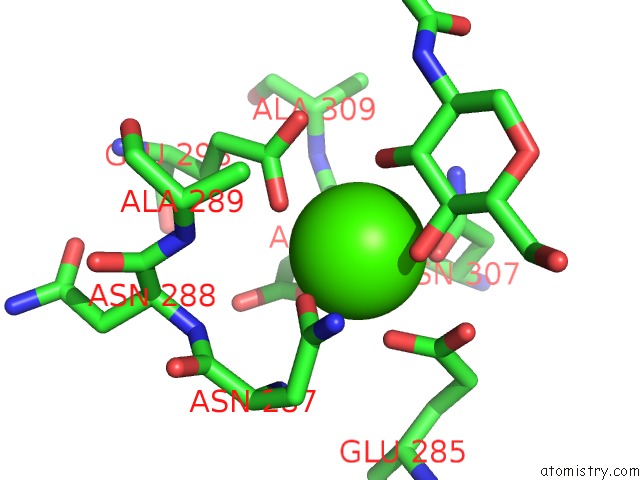
Mono view
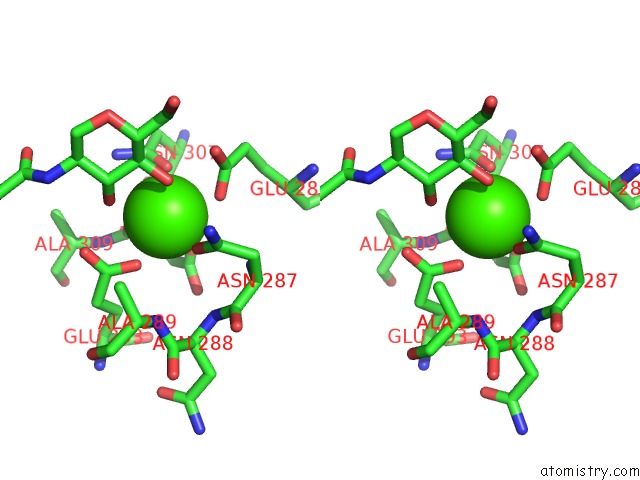
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Structure of Langerin Crd I313 Complexed with Glcnac-BETA1-3GAL-BETA1- 4GLC-Beta-CH2CH2N3 within 5.0Å range:
|
Calcium binding site 4 out of 4 in 4n35
Go back to
Calcium binding site 4 out
of 4 in the Structure of Langerin Crd I313 Complexed with Glcnac-BETA1-3GAL-BETA1- 4GLC-Beta-CH2CH2N3

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of Structure of Langerin Crd I313 Complexed with Glcnac-BETA1-3GAL-BETA1- 4GLC-Beta-CH2CH2N3 within 5.0Å range:
|
Reference:
H.Feinberg,
T.J.Rowntree,
S.L.Tan,
K.Drickamer,
W.I.Weis,
M.E.Taylor.
Common Polymorphisms in Human Langerin Change Specificity For Glycan Ligands. J.Biol.Chem. V. 288 36762 2013.
ISSN: ISSN 0021-9258
PubMed: 24217250
DOI: 10.1074/JBC.M113.528000
Page generated: Wed Jul 9 00:41:08 2025
ISSN: ISSN 0021-9258
PubMed: 24217250
DOI: 10.1074/JBC.M113.528000
Last articles
Cl in 5KOECl in 5KP1
Cl in 5KOP
Cl in 5KP3
Cl in 5KOR
Cl in 5KOL
Cl in 5KN1
Cl in 5KMU
Cl in 5KMD
Cl in 5KMB