Calcium »
PDB 4xut-4y9b »
4y7e »
Calcium in PDB 4y7e: Crystal Structure of Beta-Mannanase From Streptomyces Thermolilacinus with Mannohexaose
Enzymatic activity of Crystal Structure of Beta-Mannanase From Streptomyces Thermolilacinus with Mannohexaose
All present enzymatic activity of Crystal Structure of Beta-Mannanase From Streptomyces Thermolilacinus with Mannohexaose:
3.2.1.4;
3.2.1.4;
Protein crystallography data
The structure of Crystal Structure of Beta-Mannanase From Streptomyces Thermolilacinus with Mannohexaose, PDB code: 4y7e
was solved by
Y.Kumagai,
K.Yamashita,
M.Okuyama,
T.Hatanaka,
M.Yao,
A.Kimura,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.46 / 1.50 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 65.709, 100.706, 104.737, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.6 / 17.3 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of Beta-Mannanase From Streptomyces Thermolilacinus with Mannohexaose
(pdb code 4y7e). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 5 binding sites of Calcium where determined in the Crystal Structure of Beta-Mannanase From Streptomyces Thermolilacinus with Mannohexaose, PDB code: 4y7e:
Jump to Calcium binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Calcium where determined in the Crystal Structure of Beta-Mannanase From Streptomyces Thermolilacinus with Mannohexaose, PDB code: 4y7e:
Jump to Calcium binding site number: 1; 2; 3; 4; 5;
Calcium binding site 1 out of 5 in 4y7e
Go back to
Calcium binding site 1 out
of 5 in the Crystal Structure of Beta-Mannanase From Streptomyces Thermolilacinus with Mannohexaose
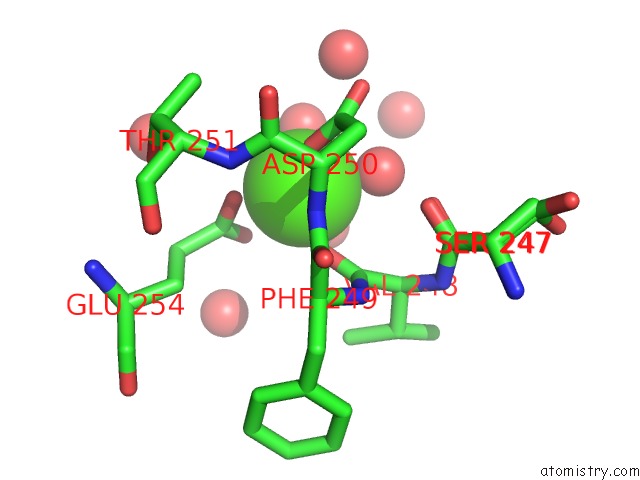
Mono view
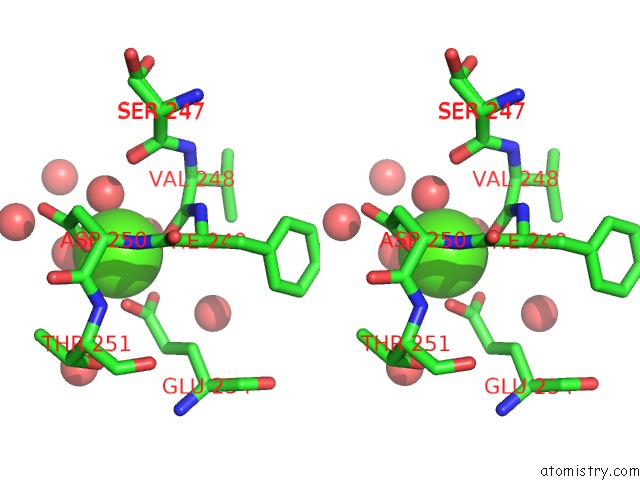
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of Beta-Mannanase From Streptomyces Thermolilacinus with Mannohexaose within 5.0Å range:
|
Calcium binding site 2 out of 5 in 4y7e
Go back to
Calcium binding site 2 out
of 5 in the Crystal Structure of Beta-Mannanase From Streptomyces Thermolilacinus with Mannohexaose
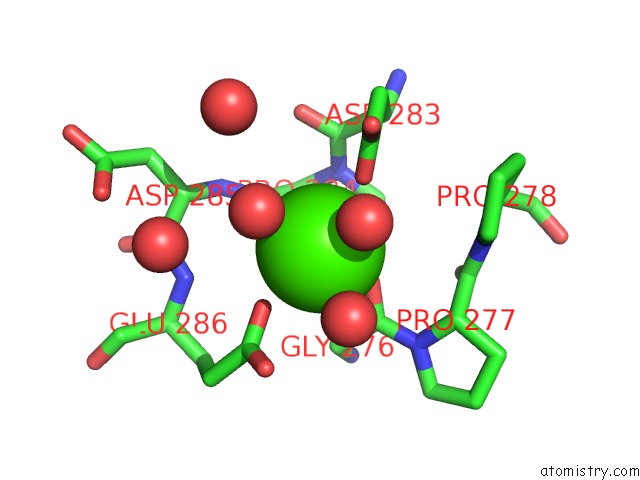
Mono view
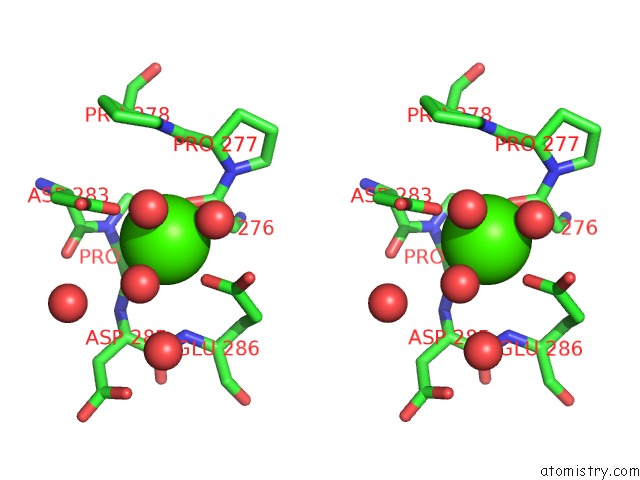
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Crystal Structure of Beta-Mannanase From Streptomyces Thermolilacinus with Mannohexaose within 5.0Å range:
|
Calcium binding site 3 out of 5 in 4y7e
Go back to
Calcium binding site 3 out
of 5 in the Crystal Structure of Beta-Mannanase From Streptomyces Thermolilacinus with Mannohexaose
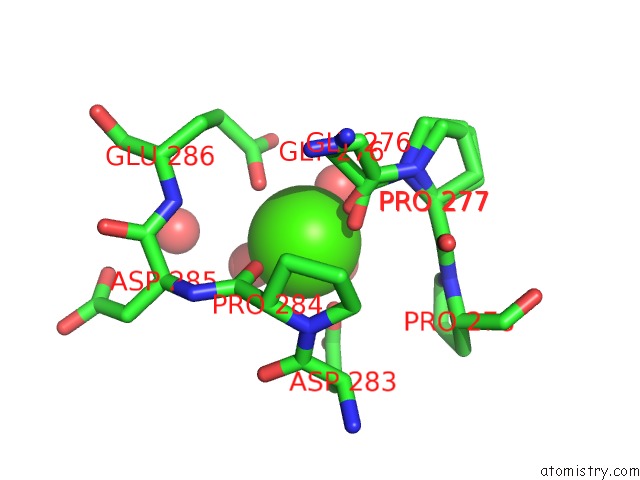
Mono view
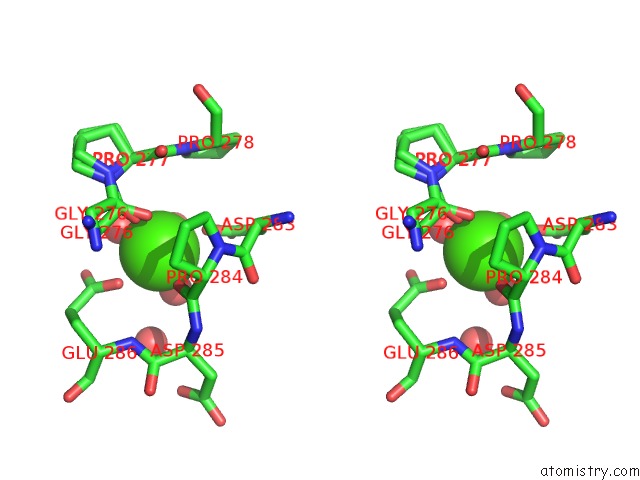
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Crystal Structure of Beta-Mannanase From Streptomyces Thermolilacinus with Mannohexaose within 5.0Å range:
|
Calcium binding site 4 out of 5 in 4y7e
Go back to
Calcium binding site 4 out
of 5 in the Crystal Structure of Beta-Mannanase From Streptomyces Thermolilacinus with Mannohexaose
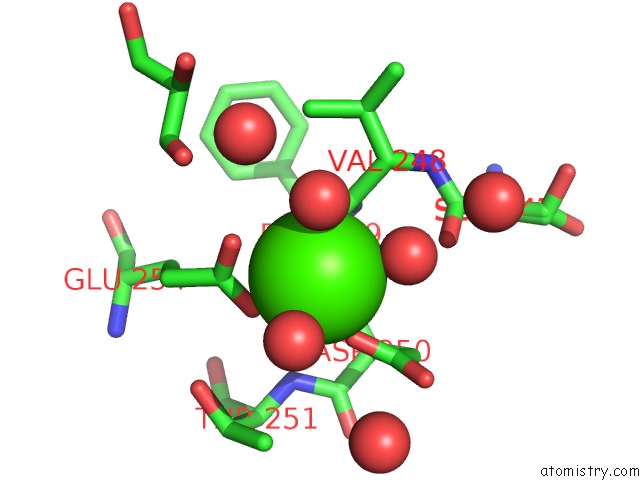
Mono view
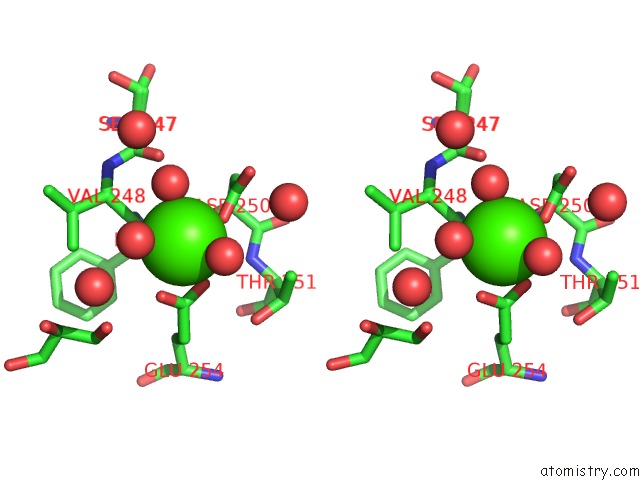
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of Crystal Structure of Beta-Mannanase From Streptomyces Thermolilacinus with Mannohexaose within 5.0Å range:
|
Calcium binding site 5 out of 5 in 4y7e
Go back to
Calcium binding site 5 out
of 5 in the Crystal Structure of Beta-Mannanase From Streptomyces Thermolilacinus with Mannohexaose
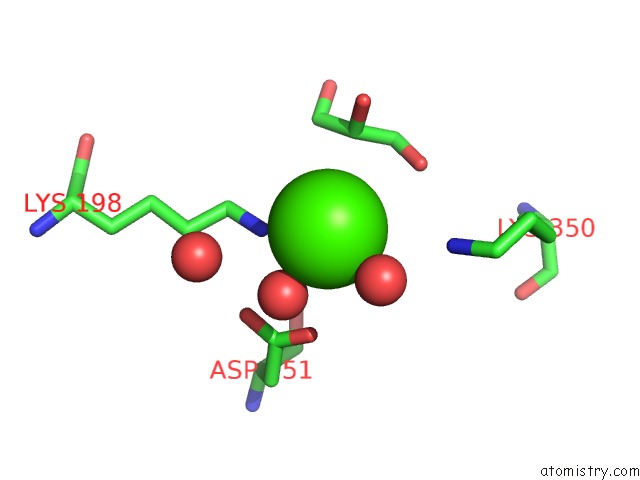
Mono view
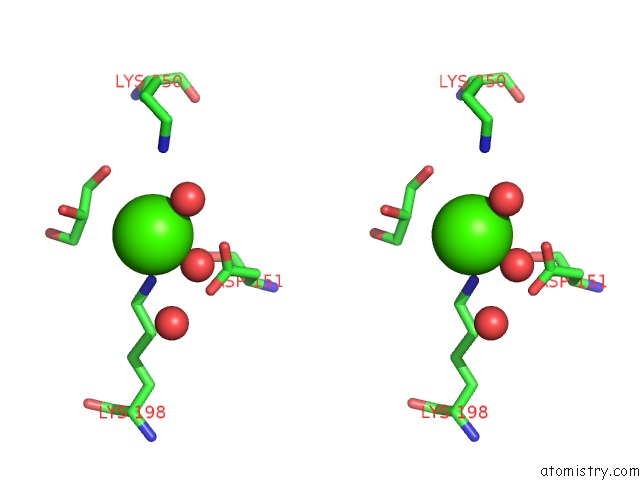
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 5 of Crystal Structure of Beta-Mannanase From Streptomyces Thermolilacinus with Mannohexaose within 5.0Å range:
|
Reference:
Y.Kumagai,
K.Yamashita,
T.Tagami,
M.Uraji,
K.Wan,
M.Okuyama,
M.Yao,
A.Kimura,
T.Hatanaka.
The Loop Structure of Actinomycete Glycoside Hydrolase Family 5 Mannanases Governs Substrate Recognition Febs J. V. 282 4001 2015.
ISSN: ISSN 1742-464X
PubMed: 26257335
DOI: 10.1111/FEBS.13401
Page generated: Wed Jul 9 03:03:39 2025
ISSN: ISSN 1742-464X
PubMed: 26257335
DOI: 10.1111/FEBS.13401
Last articles
Ca in 7JNPCa in 7JPX
Ca in 7JPW
Ca in 7JPV
Ca in 7JPL
Ca in 7JMS
Ca in 7JPK
Ca in 7JND
Ca in 7JNF
Ca in 7JNB