Calcium »
PDB 5ckm-5cxl »
5cpl »
Calcium in PDB 5cpl: The Crystal Structure of Xenobiotic Reductase A (Xena) From Pseudomonas Putida in Complex with A Nicotinamide Mimic (MNH2)
Protein crystallography data
The structure of The Crystal Structure of Xenobiotic Reductase A (Xena) From Pseudomonas Putida in Complex with A Nicotinamide Mimic (MNH2), PDB code: 5cpl
was solved by
T.Knaus,
C.E.Paul,
C.W.Levy,
F.G.Mutti,
F.Hollmann,
N.S.Scrutton,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.02 / 1.57 |
| Space group | P 2 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 56.800, 83.820, 155.880, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 13.5 / 16 |
Calcium Binding Sites:
The binding sites of Calcium atom in the The Crystal Structure of Xenobiotic Reductase A (Xena) From Pseudomonas Putida in Complex with A Nicotinamide Mimic (MNH2)
(pdb code 5cpl). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total only one binding site of Calcium was determined in the The Crystal Structure of Xenobiotic Reductase A (Xena) From Pseudomonas Putida in Complex with A Nicotinamide Mimic (MNH2), PDB code: 5cpl:
In total only one binding site of Calcium was determined in the The Crystal Structure of Xenobiotic Reductase A (Xena) From Pseudomonas Putida in Complex with A Nicotinamide Mimic (MNH2), PDB code: 5cpl:
Calcium binding site 1 out of 1 in 5cpl
Go back to
Calcium binding site 1 out
of 1 in the The Crystal Structure of Xenobiotic Reductase A (Xena) From Pseudomonas Putida in Complex with A Nicotinamide Mimic (MNH2)
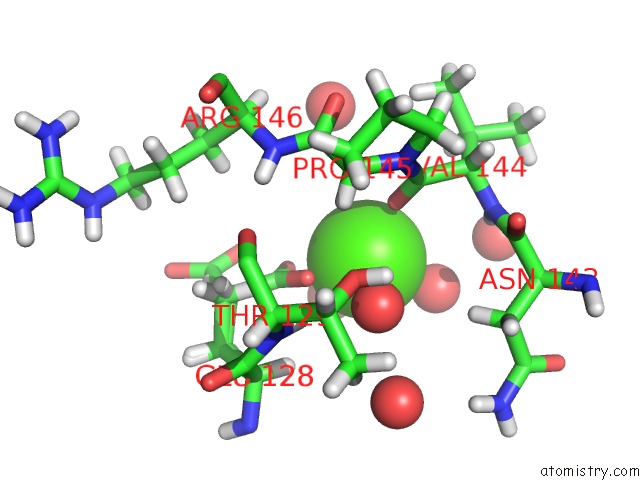
Mono view
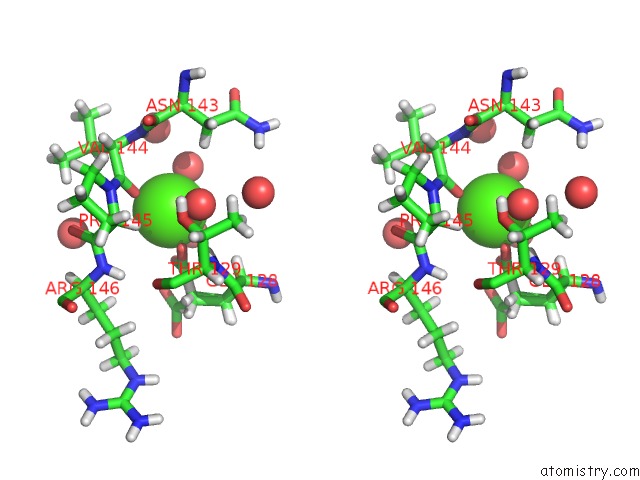
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of The Crystal Structure of Xenobiotic Reductase A (Xena) From Pseudomonas Putida in Complex with A Nicotinamide Mimic (MNH2) within 5.0Å range:
|
Reference:
T.Knaus,
C.E.Paul,
C.W.Levy,
S.De Vries,
F.G.Mutti,
F.Hollmann,
N.S.Scrutton.
Better Than Nature: Nicotinamide Biomimetics That Outperform Natural Coenzymes. J.Am.Chem.Soc. V. 138 1033 2016.
ISSN: ESSN 1520-5126
PubMed: 26727612
DOI: 10.1021/JACS.5B12252
Page generated: Wed Jul 9 04:39:02 2025
ISSN: ESSN 1520-5126
PubMed: 26727612
DOI: 10.1021/JACS.5B12252
Last articles
F in 7N2AF in 7MXY
F in 7MYY
F in 7N13
F in 7MYU
F in 7MYR
F in 7MYO
F in 7MXN
F in 7MXG
F in 7MXH