Calcium »
PDB 5npf-5odc »
5nrm »
Calcium in PDB 5nrm: Crystal Structure of the Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B in Complex with CEL5 Dockerin S51I, L52N Mutant
Enzymatic activity of Crystal Structure of the Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B in Complex with CEL5 Dockerin S51I, L52N Mutant
All present enzymatic activity of Crystal Structure of the Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B in Complex with CEL5 Dockerin S51I, L52N Mutant:
3.2.1.4;
3.2.1.4;
Protein crystallography data
The structure of Crystal Structure of the Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B in Complex with CEL5 Dockerin S51I, L52N Mutant, PDB code: 5nrm
was solved by
P.Bule,
S.Najmudin,
C.M.G.A.Fontes,
V.D.Alves,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 49.05 / 1.40 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 30.490, 59.950, 51.260, 90.00, 106.88, 90.00 |
| R / Rfree (%) | 17.8 / 20.5 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of the Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B in Complex with CEL5 Dockerin S51I, L52N Mutant
(pdb code 5nrm). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 3 binding sites of Calcium where determined in the Crystal Structure of the Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B in Complex with CEL5 Dockerin S51I, L52N Mutant, PDB code: 5nrm:
Jump to Calcium binding site number: 1; 2; 3;
In total 3 binding sites of Calcium where determined in the Crystal Structure of the Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B in Complex with CEL5 Dockerin S51I, L52N Mutant, PDB code: 5nrm:
Jump to Calcium binding site number: 1; 2; 3;
Calcium binding site 1 out of 3 in 5nrm
Go back to
Calcium binding site 1 out
of 3 in the Crystal Structure of the Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B in Complex with CEL5 Dockerin S51I, L52N Mutant
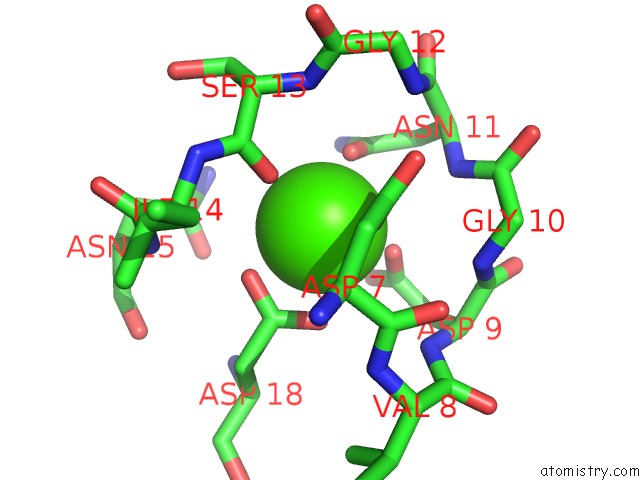
Mono view
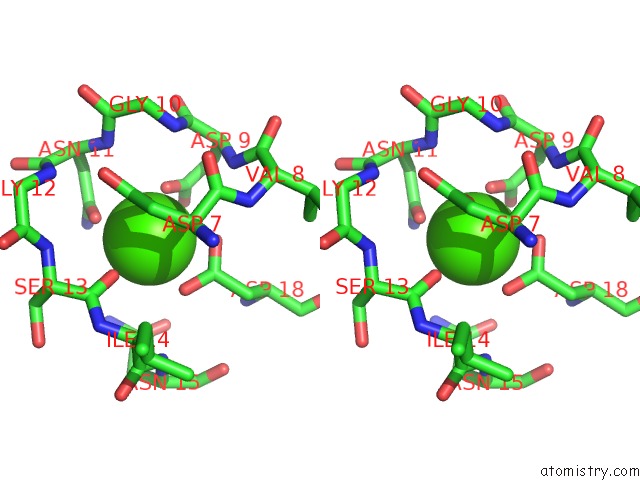
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of the Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B in Complex with CEL5 Dockerin S51I, L52N Mutant within 5.0Å range:
|
Calcium binding site 2 out of 3 in 5nrm
Go back to
Calcium binding site 2 out
of 3 in the Crystal Structure of the Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B in Complex with CEL5 Dockerin S51I, L52N Mutant
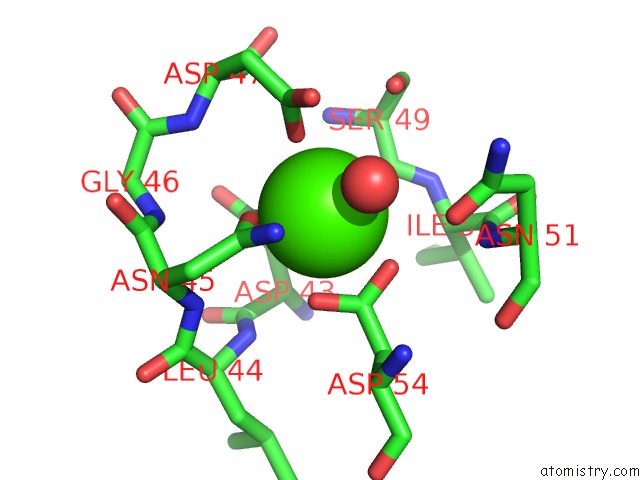
Mono view
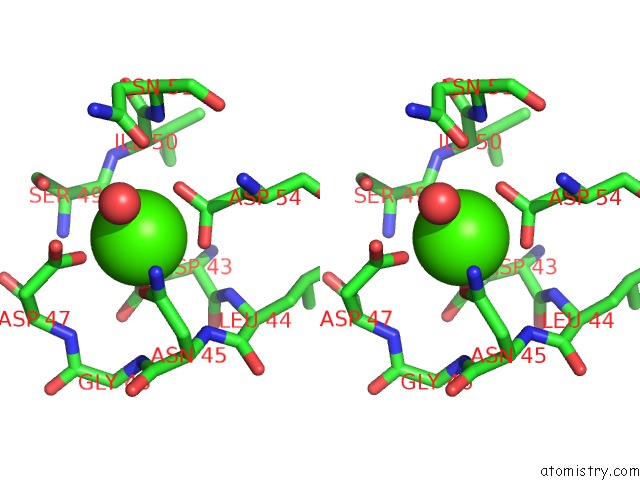
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Crystal Structure of the Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B in Complex with CEL5 Dockerin S51I, L52N Mutant within 5.0Å range:
|
Calcium binding site 3 out of 3 in 5nrm
Go back to
Calcium binding site 3 out
of 3 in the Crystal Structure of the Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B in Complex with CEL5 Dockerin S51I, L52N Mutant
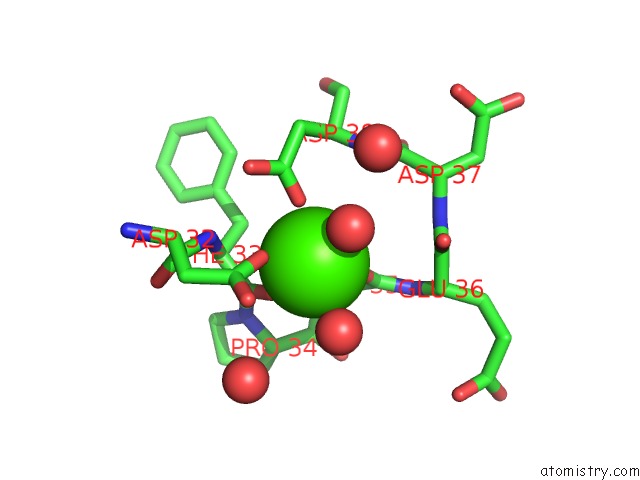
Mono view
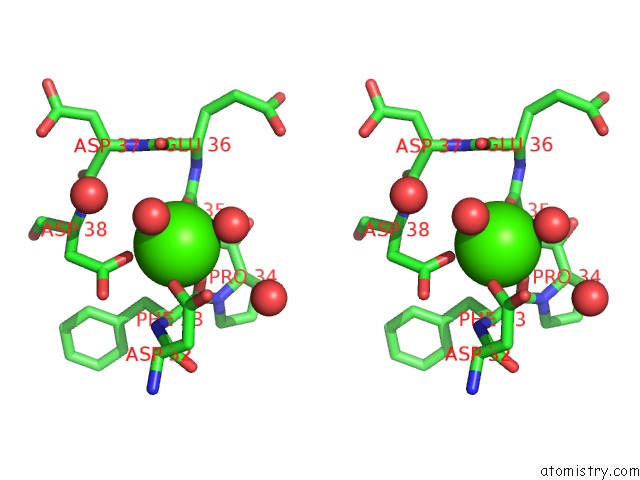
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Crystal Structure of the Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B in Complex with CEL5 Dockerin S51I, L52N Mutant within 5.0Å range:
|
Reference:
P.Bule,
K.Cameron,
J.A.M.Prates,
L.M.A.Ferreira,
S.P.Smith,
H.J.Gilbert,
E.A.Bayer,
S.Najmudin,
C.M.G.A.Fontes,
V.D.Alves.
Structure-Function Analyses Generate Novel Specificities to Assemble the Components of Multienzyme Bacterial Cellulosome Complexes. J. Biol. Chem. V. 293 4201 2018.
ISSN: ESSN 1083-351X
PubMed: 29367338
DOI: 10.1074/JBC.RA117.001241
Page generated: Wed Jul 9 09:08:33 2025
ISSN: ESSN 1083-351X
PubMed: 29367338
DOI: 10.1074/JBC.RA117.001241
Last articles
F in 7ORZF in 7OP1
F in 7ORF
F in 7OP8
F in 7ORE
F in 7OP5
F in 7OL8
F in 7OL7
F in 7OL3
F in 7OOB