Calcium »
PDB 5ufq-5uw6 »
5uu7 »
Calcium in PDB 5uu7: Tetragonal Thermolysin (295 K) in the Presence of 50% Mpd
Enzymatic activity of Tetragonal Thermolysin (295 K) in the Presence of 50% Mpd
All present enzymatic activity of Tetragonal Thermolysin (295 K) in the Presence of 50% Mpd:
3.4.24.27;
3.4.24.27;
Protein crystallography data
The structure of Tetragonal Thermolysin (295 K) in the Presence of 50% Mpd, PDB code: 5uu7
was solved by
D.H.Juers,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.76 / 1.60 |
| Space group | P 41 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 98.026, 98.026, 107.522, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.4 / 15.6 |
Other elements in 5uu7:
The structure of Tetragonal Thermolysin (295 K) in the Presence of 50% Mpd also contains other interesting chemical elements:
| Zinc | (Zn) | 8 atoms |
| Chlorine | (Cl) | 4 atoms |
Calcium Binding Sites:
The binding sites of Calcium atom in the Tetragonal Thermolysin (295 K) in the Presence of 50% Mpd
(pdb code 5uu7). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 3 binding sites of Calcium where determined in the Tetragonal Thermolysin (295 K) in the Presence of 50% Mpd, PDB code: 5uu7:
Jump to Calcium binding site number: 1; 2; 3;
In total 3 binding sites of Calcium where determined in the Tetragonal Thermolysin (295 K) in the Presence of 50% Mpd, PDB code: 5uu7:
Jump to Calcium binding site number: 1; 2; 3;
Calcium binding site 1 out of 3 in 5uu7
Go back to
Calcium binding site 1 out
of 3 in the Tetragonal Thermolysin (295 K) in the Presence of 50% Mpd
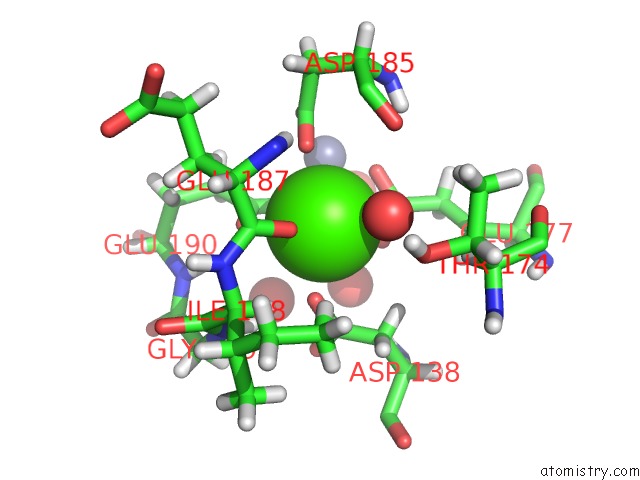
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Tetragonal Thermolysin (295 K) in the Presence of 50% Mpd within 5.0Å range:
|
Calcium binding site 2 out of 3 in 5uu7
Go back to
Calcium binding site 2 out
of 3 in the Tetragonal Thermolysin (295 K) in the Presence of 50% Mpd

Mono view
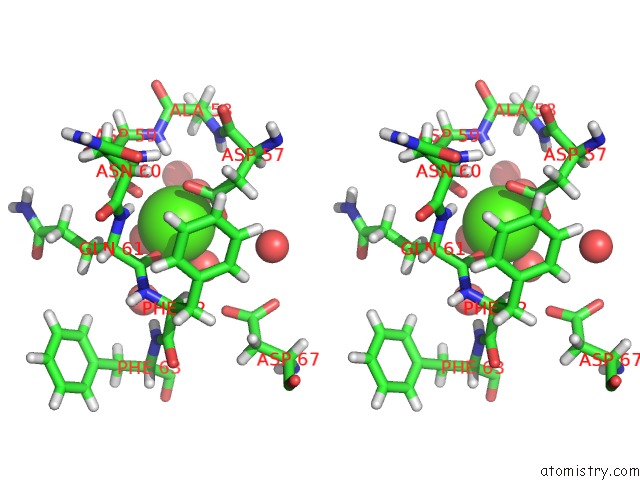
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Tetragonal Thermolysin (295 K) in the Presence of 50% Mpd within 5.0Å range:
|
Calcium binding site 3 out of 3 in 5uu7
Go back to
Calcium binding site 3 out
of 3 in the Tetragonal Thermolysin (295 K) in the Presence of 50% Mpd

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Tetragonal Thermolysin (295 K) in the Presence of 50% Mpd within 5.0Å range:
|
Reference:
D.H.Juers,
C.A.Farley,
C.P.Saxby,
R.A.Cotter,
J.K.B.Cahn,
R.C.Holton-Burke,
K.Harrison,
Z.Wu.
The Impact of Cryosolution Thermal Contraction on Proteins and Protein Crystals: Volumes, Conformation and Order. Acta Crystallogr D Struct V. 74 922 2018BIOL.
ISSN: ISSN 2059-7983
PubMed: 30198901
DOI: 10.1107/S2059798318008793
Page generated: Wed Jul 9 10:34:34 2025
ISSN: ISSN 2059-7983
PubMed: 30198901
DOI: 10.1107/S2059798318008793
Last articles
F in 7MLDF in 7MKX
F in 7MGK
F in 7MGJ
F in 7MHD
F in 7MFH
F in 7MHC
F in 7MGE
F in 7MFD
F in 7MEW