Calcium »
PDB 5xkf-5xu9 »
5xqo »
Calcium in PDB 5xqo: Crystal Structure of A Pl 26 Exo-Rhamnogalacturonan Lyase From Penicillium Chrysogenum Complexed with Tetrameric Substrate
Protein crystallography data
The structure of Crystal Structure of A Pl 26 Exo-Rhamnogalacturonan Lyase From Penicillium Chrysogenum Complexed with Tetrameric Substrate, PDB code: 5xqo
was solved by
Y.Kunishige,
M.Iwai,
T.Tada,
S.Nishimura,
T.Sakamoto,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 119.58 / 3.20 |
| Space group | P 42 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 166.437, 166.437, 171.932, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.1 / 26.4 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of A Pl 26 Exo-Rhamnogalacturonan Lyase From Penicillium Chrysogenum Complexed with Tetrameric Substrate
(pdb code 5xqo). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Crystal Structure of A Pl 26 Exo-Rhamnogalacturonan Lyase From Penicillium Chrysogenum Complexed with Tetrameric Substrate, PDB code: 5xqo:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Crystal Structure of A Pl 26 Exo-Rhamnogalacturonan Lyase From Penicillium Chrysogenum Complexed with Tetrameric Substrate, PDB code: 5xqo:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 5xqo
Go back to
Calcium binding site 1 out
of 2 in the Crystal Structure of A Pl 26 Exo-Rhamnogalacturonan Lyase From Penicillium Chrysogenum Complexed with Tetrameric Substrate
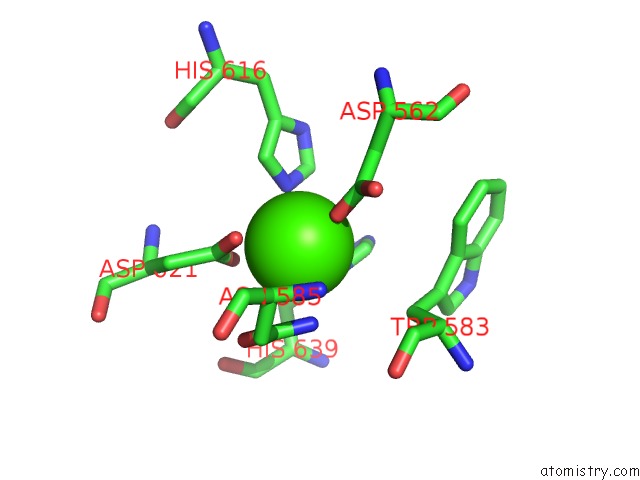
Mono view
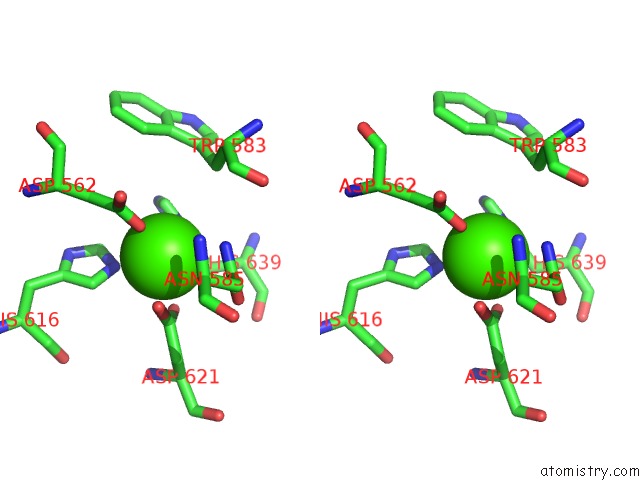
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of A Pl 26 Exo-Rhamnogalacturonan Lyase From Penicillium Chrysogenum Complexed with Tetrameric Substrate within 5.0Å range:
|
Calcium binding site 2 out of 2 in 5xqo
Go back to
Calcium binding site 2 out
of 2 in the Crystal Structure of A Pl 26 Exo-Rhamnogalacturonan Lyase From Penicillium Chrysogenum Complexed with Tetrameric Substrate
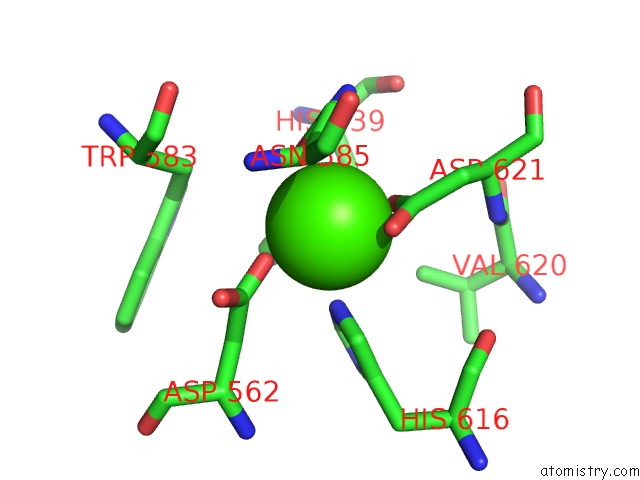
Mono view
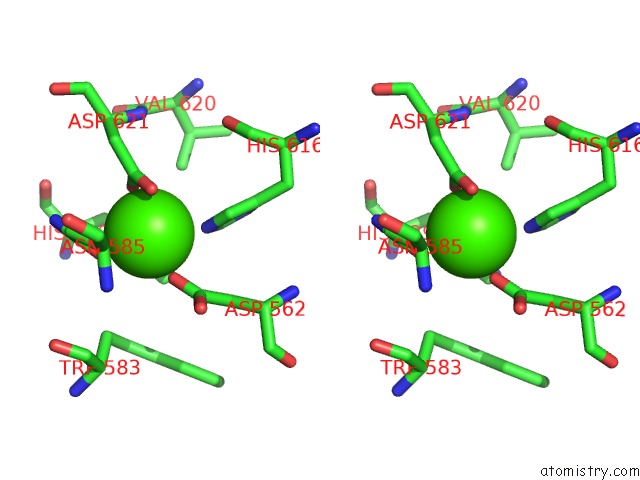
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Crystal Structure of A Pl 26 Exo-Rhamnogalacturonan Lyase From Penicillium Chrysogenum Complexed with Tetrameric Substrate within 5.0Å range:
|
Reference:
Y.Kunishige,
M.Iwai,
M.Nakazawa,
M.Ueda,
T.Tada,
S.Nishimura,
T.Sakamoto.
Crystal Structure of Exo-Rhamnogalacturonan Lyase From Penicillium Chrysogenum As A Member of Polysaccharide Lyase Family 26 Febs Lett. V. 592 1378 2018.
ISSN: ISSN 1873-3468
PubMed: 29574769
DOI: 10.1002/1873-3468.13034
Page generated: Wed Jul 9 11:45:34 2025
ISSN: ISSN 1873-3468
PubMed: 29574769
DOI: 10.1002/1873-3468.13034
Last articles
Ca in 7NSDCa in 7NQC
Ca in 7NMI
Ca in 7NP8
Ca in 7NSA
Ca in 7NS9
Ca in 7NRK
Ca in 7NPG
Ca in 7NPB
Ca in 7NN9