Calcium »
PDB 6b5v-6bia »
6b6n »
Calcium in PDB 6b6n: Orthorhombic Trypsin (295 K) in the Presence of 50% Mpd
Enzymatic activity of Orthorhombic Trypsin (295 K) in the Presence of 50% Mpd
All present enzymatic activity of Orthorhombic Trypsin (295 K) in the Presence of 50% Mpd:
3.4.21.4;
3.4.21.4;
Protein crystallography data
The structure of Orthorhombic Trypsin (295 K) in the Presence of 50% Mpd, PDB code: 6b6n
was solved by
D.H.Juers,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 13.17 / 2.00 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 54.692, 58.538, 67.576, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 12.1 / 15.7 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Orthorhombic Trypsin (295 K) in the Presence of 50% Mpd
(pdb code 6b6n). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total only one binding site of Calcium was determined in the Orthorhombic Trypsin (295 K) in the Presence of 50% Mpd, PDB code: 6b6n:
In total only one binding site of Calcium was determined in the Orthorhombic Trypsin (295 K) in the Presence of 50% Mpd, PDB code: 6b6n:
Calcium binding site 1 out of 1 in 6b6n
Go back to
Calcium binding site 1 out
of 1 in the Orthorhombic Trypsin (295 K) in the Presence of 50% Mpd
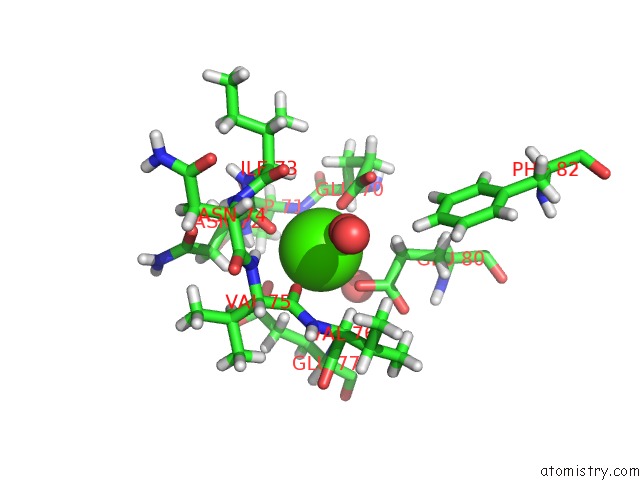
Mono view
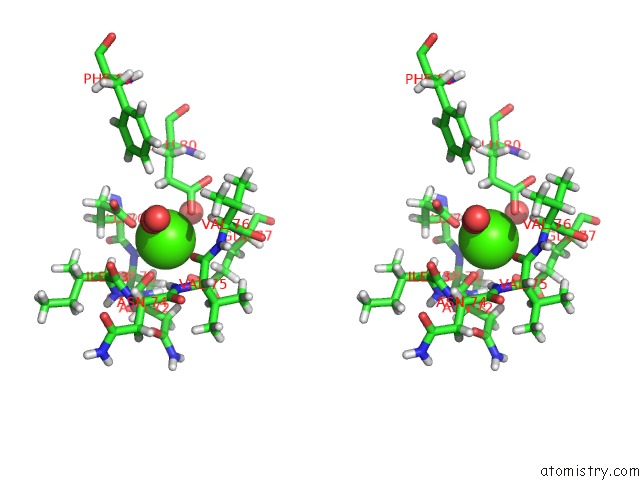
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Orthorhombic Trypsin (295 K) in the Presence of 50% Mpd within 5.0Å range:
|
Reference:
D.H.Juers,
C.A.Farley,
C.P.Saxby,
R.A.Cotter,
J.K.B.Cahn,
R.C.Holton-Burke,
K.Harrison,
Z.Wu.
The Impact of Cryosolution Thermal Contraction on Proteins and Protein Crystals: Volumes, Conformation and Order. Acta Crystallogr D Struct V. 74 922 2018BIOL.
ISSN: ISSN 2059-7983
PubMed: 30198901
DOI: 10.1107/S2059798318008793
Page generated: Wed Jul 9 12:36:40 2025
ISSN: ISSN 2059-7983
PubMed: 30198901
DOI: 10.1107/S2059798318008793
Last articles
F in 7MLDF in 7MKX
F in 7MGK
F in 7MGJ
F in 7MHD
F in 7MFH
F in 7MHC
F in 7MGE
F in 7MFD
F in 7MEW