Calcium »
PDB 6hi0-6i1q »
6htv »
Calcium in PDB 6htv: Crystal Structure of Leuconostoc Citreum Nrrl B-1299 N-Terminally Truncated Dextransucrase Dsr-M in Complex with Isomaltotetraose
Enzymatic activity of Crystal Structure of Leuconostoc Citreum Nrrl B-1299 N-Terminally Truncated Dextransucrase Dsr-M in Complex with Isomaltotetraose
All present enzymatic activity of Crystal Structure of Leuconostoc Citreum Nrrl B-1299 N-Terminally Truncated Dextransucrase Dsr-M in Complex with Isomaltotetraose:
2.4.1.140;
2.4.1.140;
Protein crystallography data
The structure of Crystal Structure of Leuconostoc Citreum Nrrl B-1299 N-Terminally Truncated Dextransucrase Dsr-M in Complex with Isomaltotetraose, PDB code: 6htv
was solved by
M.Claverie,
G.Cioci,
M.Remaud-Simeon,
C.Moulis,
G.Lippens,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.96 / 3.90 |
| Space group | P 31 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 185.308, 185.308, 150.774, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 21 / 23.2 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of Leuconostoc Citreum Nrrl B-1299 N-Terminally Truncated Dextransucrase Dsr-M in Complex with Isomaltotetraose
(pdb code 6htv). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total only one binding site of Calcium was determined in the Crystal Structure of Leuconostoc Citreum Nrrl B-1299 N-Terminally Truncated Dextransucrase Dsr-M in Complex with Isomaltotetraose, PDB code: 6htv:
In total only one binding site of Calcium was determined in the Crystal Structure of Leuconostoc Citreum Nrrl B-1299 N-Terminally Truncated Dextransucrase Dsr-M in Complex with Isomaltotetraose, PDB code: 6htv:
Calcium binding site 1 out of 1 in 6htv
Go back to
Calcium binding site 1 out
of 1 in the Crystal Structure of Leuconostoc Citreum Nrrl B-1299 N-Terminally Truncated Dextransucrase Dsr-M in Complex with Isomaltotetraose
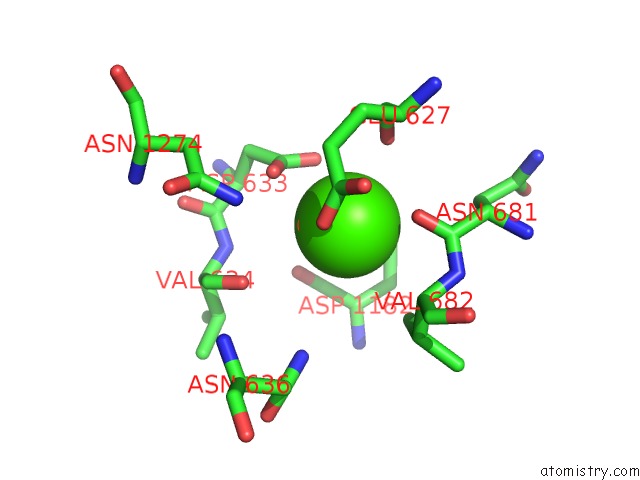
Mono view
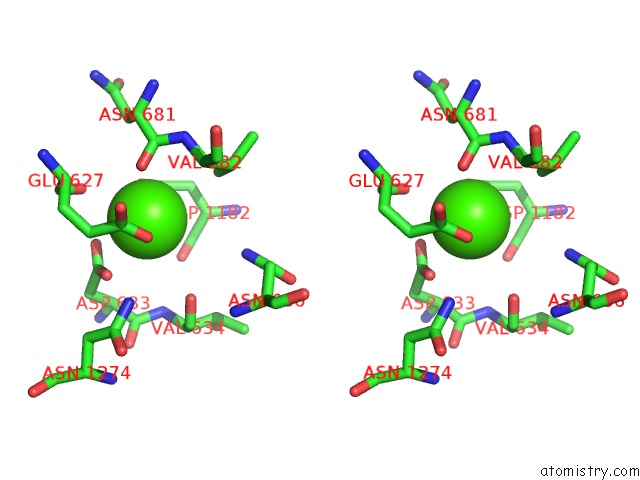
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of Leuconostoc Citreum Nrrl B-1299 N-Terminally Truncated Dextransucrase Dsr-M in Complex with Isomaltotetraose within 5.0Å range:
|
Reference:
M.Claverie,
G.Cioci,
M.Guionnet,
J.Schorghuber,
R.Lichtenecker,
C.Moulis,
M.Remaud-Simeon,
G.Lippens.
Futile Encounter Engineering of the Dsr-M Dextransucrase Modifies the Resulting Polymer Length. Biochemistry V. 58 2853 2019.
ISSN: ISSN 0006-2960
PubMed: 31140266
DOI: 10.1021/ACS.BIOCHEM.9B00373
Page generated: Wed Jul 9 14:42:41 2025
ISSN: ISSN 0006-2960
PubMed: 31140266
DOI: 10.1021/ACS.BIOCHEM.9B00373
Last articles
Ca in 7LSACa in 7LRX
Ca in 7LRM
Ca in 7LQC
Ca in 7LRI
Ca in 7LQB
Ca in 7LQA
Ca in 7LRF
Ca in 7LQ9
Ca in 7LQ8