Calcium »
PDB 6oly-6p8x »
6p73 »
Calcium in PDB 6p73: Cytochrome-C-Nitrite Reductase
Enzymatic activity of Cytochrome-C-Nitrite Reductase
All present enzymatic activity of Cytochrome-C-Nitrite Reductase:
1.7.2.2;
1.7.2.2;
Protein crystallography data
The structure of Cytochrome-C-Nitrite Reductase, PDB code: 6p73
was solved by
M.Schmidt,
A.Pacheco,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.78 / 1.65 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 50.180, 96.070, 222.600, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.1 / 21.2 |
Other elements in 6p73:
The structure of Cytochrome-C-Nitrite Reductase also contains other interesting chemical elements:
| Iron | (Fe) | 10 atoms |
Calcium Binding Sites:
The binding sites of Calcium atom in the Cytochrome-C-Nitrite Reductase
(pdb code 6p73). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Cytochrome-C-Nitrite Reductase, PDB code: 6p73:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Cytochrome-C-Nitrite Reductase, PDB code: 6p73:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 6p73
Go back to
Calcium binding site 1 out
of 2 in the Cytochrome-C-Nitrite Reductase
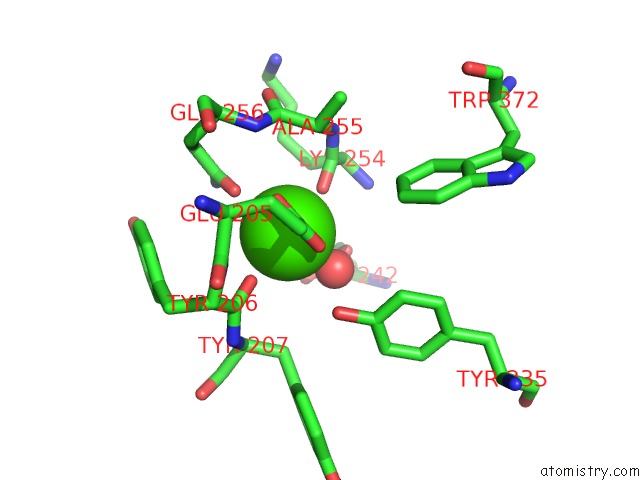
Mono view
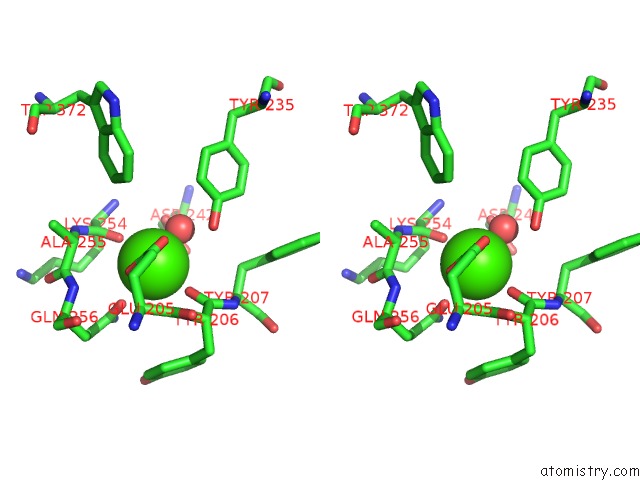
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Cytochrome-C-Nitrite Reductase within 5.0Å range:
|
Calcium binding site 2 out of 2 in 6p73
Go back to
Calcium binding site 2 out
of 2 in the Cytochrome-C-Nitrite Reductase
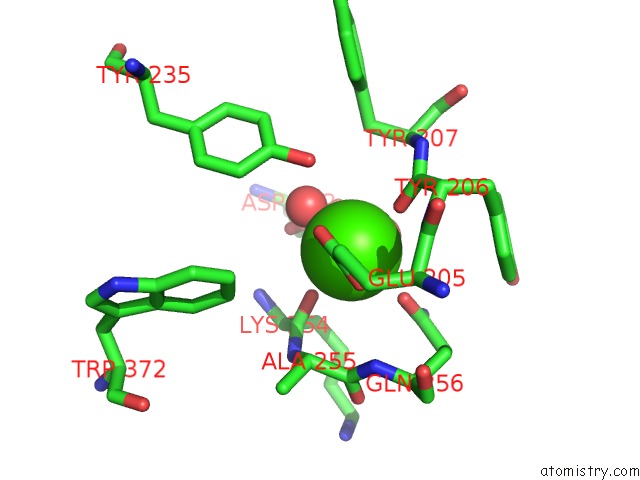
Mono view
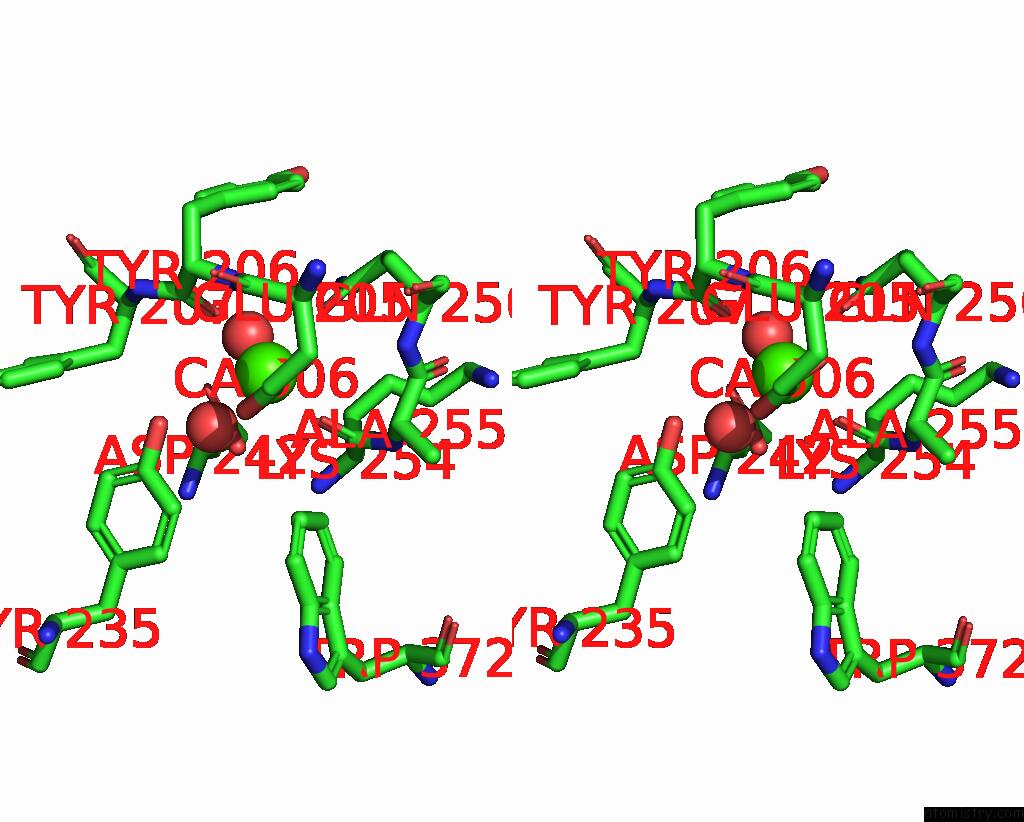
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Cytochrome-C-Nitrite Reductase within 5.0Å range:
|
Reference:
M.Ali,
N.Stein,
Y.Mao,
S.Shahid,
M.Schmidt,
B.Bennett,
A.A.Pacheco.
Trapping of A Putative Intermediate in the Cytochromecnitrite Reductase (Ccnir)-Catalyzed Reduction of Nitrite: Implications For the Ccnir Reaction Mechanism. J.Am.Chem.Soc. V. 141 13358 2019.
ISSN: ESSN 1520-5126
PubMed: 31381304
DOI: 10.1021/JACS.9B03036
Page generated: Wed Jul 9 16:43:20 2025
ISSN: ESSN 1520-5126
PubMed: 31381304
DOI: 10.1021/JACS.9B03036
Last articles
Cl in 8EN9Cl in 8EJN
Cl in 8ELA
Cl in 8EL0
Cl in 8EKX
Cl in 8EJV
Cl in 8ECY
Cl in 8EG6
Cl in 8EFL
Cl in 8EJM