Calcium »
PDB 6v4s-6vgf »
6vc3 »
Calcium in PDB 6vc3: Peanut Lectin Complexed with S-Beta-D-Thiogalactopyranosyl 6-Deoxy-6- S-Propynyl-Beta-D-Glucopyranoside (Stg)
Protein crystallography data
The structure of Peanut Lectin Complexed with S-Beta-D-Thiogalactopyranosyl 6-Deoxy-6- S-Propynyl-Beta-D-Glucopyranoside (Stg), PDB code: 6vc3
was solved by
L.H.Otero,
E.D.Primo,
A.J.Cagnoni,
M.E.Cano,
S.Klinke,
F.A.Goldbaum,
M.L.Uhrig,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 49.03 / 1.95 |
| Space group | P 2 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 76.190, 125.418, 128.112, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 26.7 / 28.3 |
Other elements in 6vc3:
The structure of Peanut Lectin Complexed with S-Beta-D-Thiogalactopyranosyl 6-Deoxy-6- S-Propynyl-Beta-D-Glucopyranoside (Stg) also contains other interesting chemical elements:
| Manganese | (Mn) | 4 atoms |
Calcium Binding Sites:
The binding sites of Calcium atom in the Peanut Lectin Complexed with S-Beta-D-Thiogalactopyranosyl 6-Deoxy-6- S-Propynyl-Beta-D-Glucopyranoside (Stg)
(pdb code 6vc3). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 4 binding sites of Calcium where determined in the Peanut Lectin Complexed with S-Beta-D-Thiogalactopyranosyl 6-Deoxy-6- S-Propynyl-Beta-D-Glucopyranoside (Stg), PDB code: 6vc3:
Jump to Calcium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Calcium where determined in the Peanut Lectin Complexed with S-Beta-D-Thiogalactopyranosyl 6-Deoxy-6- S-Propynyl-Beta-D-Glucopyranoside (Stg), PDB code: 6vc3:
Jump to Calcium binding site number: 1; 2; 3; 4;
Calcium binding site 1 out of 4 in 6vc3
Go back to
Calcium binding site 1 out
of 4 in the Peanut Lectin Complexed with S-Beta-D-Thiogalactopyranosyl 6-Deoxy-6- S-Propynyl-Beta-D-Glucopyranoside (Stg)
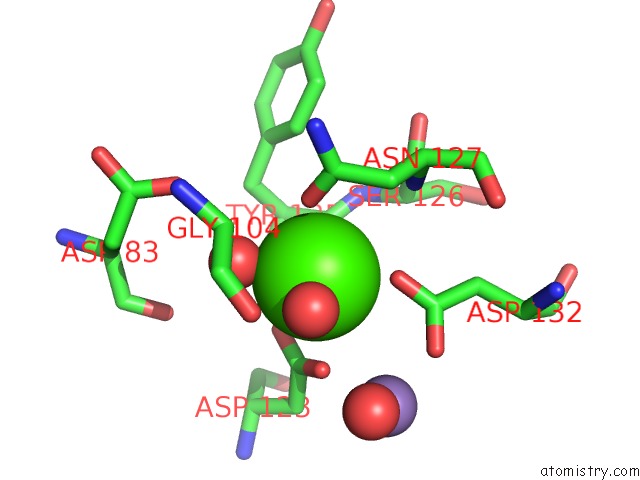
Mono view
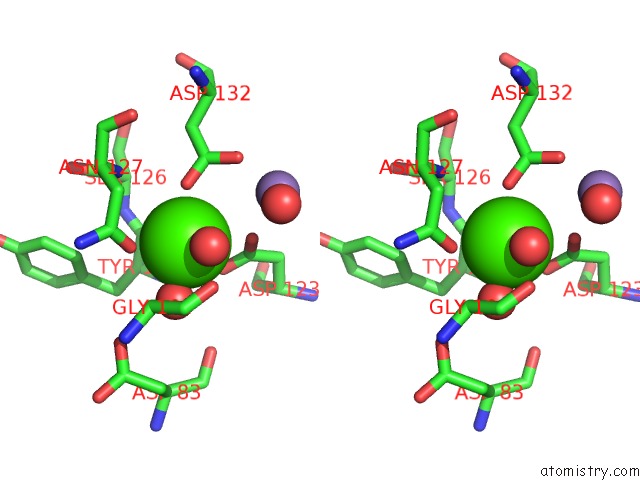
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Peanut Lectin Complexed with S-Beta-D-Thiogalactopyranosyl 6-Deoxy-6- S-Propynyl-Beta-D-Glucopyranoside (Stg) within 5.0Å range:
|
Calcium binding site 2 out of 4 in 6vc3
Go back to
Calcium binding site 2 out
of 4 in the Peanut Lectin Complexed with S-Beta-D-Thiogalactopyranosyl 6-Deoxy-6- S-Propynyl-Beta-D-Glucopyranoside (Stg)
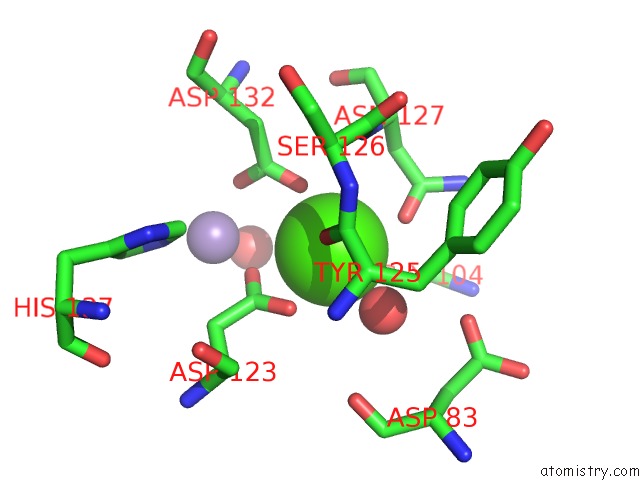
Mono view
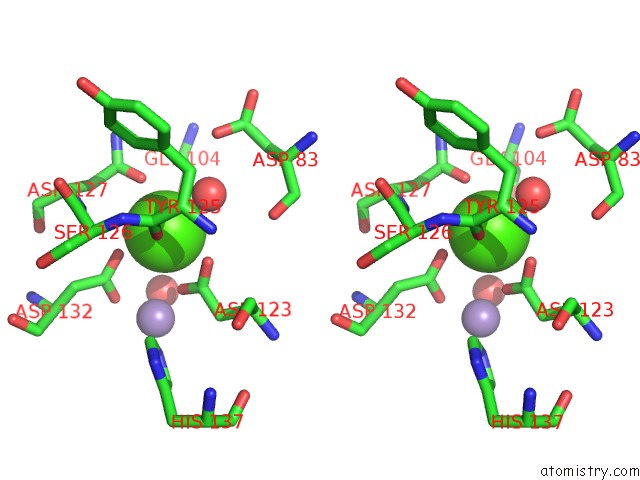
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Peanut Lectin Complexed with S-Beta-D-Thiogalactopyranosyl 6-Deoxy-6- S-Propynyl-Beta-D-Glucopyranoside (Stg) within 5.0Å range:
|
Calcium binding site 3 out of 4 in 6vc3
Go back to
Calcium binding site 3 out
of 4 in the Peanut Lectin Complexed with S-Beta-D-Thiogalactopyranosyl 6-Deoxy-6- S-Propynyl-Beta-D-Glucopyranoside (Stg)
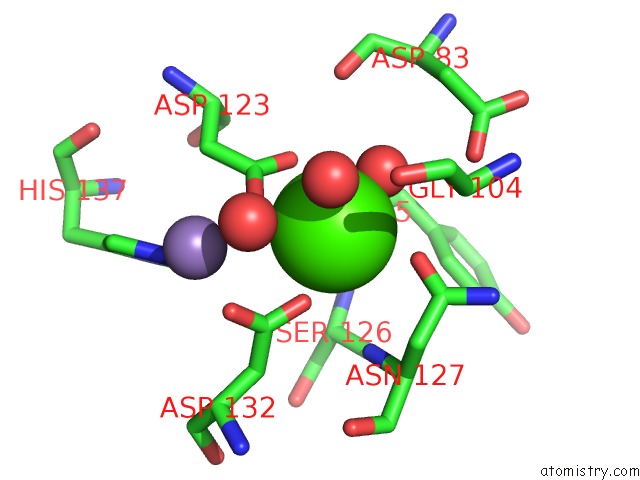
Mono view
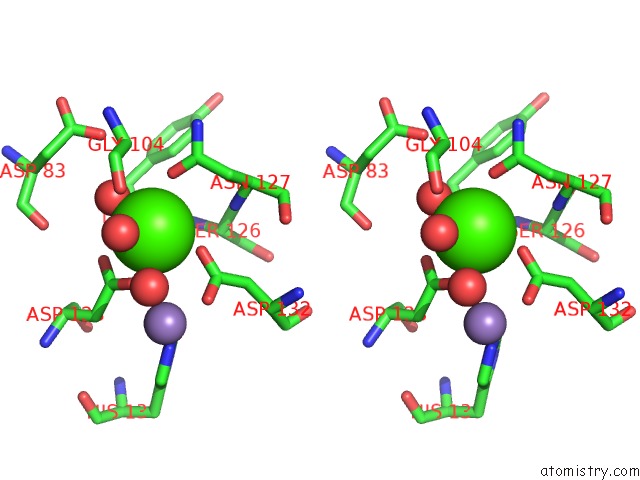
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Peanut Lectin Complexed with S-Beta-D-Thiogalactopyranosyl 6-Deoxy-6- S-Propynyl-Beta-D-Glucopyranoside (Stg) within 5.0Å range:
|
Calcium binding site 4 out of 4 in 6vc3
Go back to
Calcium binding site 4 out
of 4 in the Peanut Lectin Complexed with S-Beta-D-Thiogalactopyranosyl 6-Deoxy-6- S-Propynyl-Beta-D-Glucopyranoside (Stg)
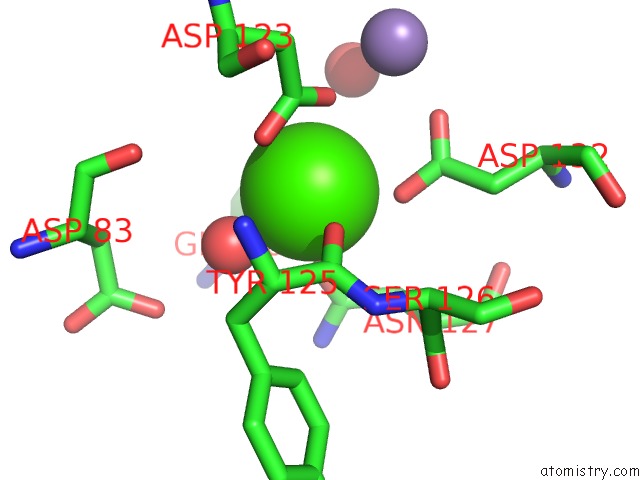
Mono view
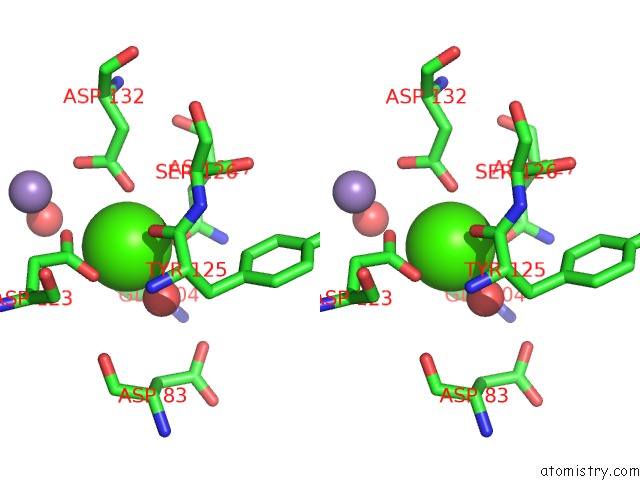
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of Peanut Lectin Complexed with S-Beta-D-Thiogalactopyranosyl 6-Deoxy-6- S-Propynyl-Beta-D-Glucopyranoside (Stg) within 5.0Å range:
|
Reference:
A.J.Cagnoni,
E.D.Primo,
S.Klinke,
M.E.Cano,
W.Giordano,
K.V.Marino,
J.Kovensky,
F.A.Goldbaum,
M.L.Uhrig,
L.H.Otero.
Crystal Structures of Peanut Lectin in the Presence of Synthetic Beta-N- and Beta-S-Galactosides Disclose Evidence For the Recognition of Different Glycomimetic Ligands. Acta Crystallogr D Struct V. 76 1080 2020BIOL.
ISSN: ISSN 2059-7983
PubMed: 33135679
DOI: 10.1107/S2059798320012371
Page generated: Wed Jul 9 19:03:36 2025
ISSN: ISSN 2059-7983
PubMed: 33135679
DOI: 10.1107/S2059798320012371
Last articles
Cl in 5QFZCl in 5QGV
Cl in 5QGS
Cl in 5QFJ
Cl in 5QF2
Cl in 5QF7
Cl in 5QED
Cl in 5QFA
Cl in 5QEW
Cl in 5QEJ