Calcium »
PDB 6yxh-6zqx »
6zhj »
Calcium in PDB 6zhj: 3D Electron Diffraction Structure of Thermolysin From Bacillus Thermoproteolyticus
Enzymatic activity of 3D Electron Diffraction Structure of Thermolysin From Bacillus Thermoproteolyticus
All present enzymatic activity of 3D Electron Diffraction Structure of Thermolysin From Bacillus Thermoproteolyticus:
3.4.24.27;
3.4.24.27;
Other elements in 6zhj:
The structure of 3D Electron Diffraction Structure of Thermolysin From Bacillus Thermoproteolyticus also contains other interesting chemical elements:
| Zinc | (Zn) | 1 atom |
Calcium Binding Sites:
The binding sites of Calcium atom in the 3D Electron Diffraction Structure of Thermolysin From Bacillus Thermoproteolyticus
(pdb code 6zhj). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 4 binding sites of Calcium where determined in the 3D Electron Diffraction Structure of Thermolysin From Bacillus Thermoproteolyticus, PDB code: 6zhj:
Jump to Calcium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Calcium where determined in the 3D Electron Diffraction Structure of Thermolysin From Bacillus Thermoproteolyticus, PDB code: 6zhj:
Jump to Calcium binding site number: 1; 2; 3; 4;
Calcium binding site 1 out of 4 in 6zhj
Go back to
Calcium binding site 1 out
of 4 in the 3D Electron Diffraction Structure of Thermolysin From Bacillus Thermoproteolyticus
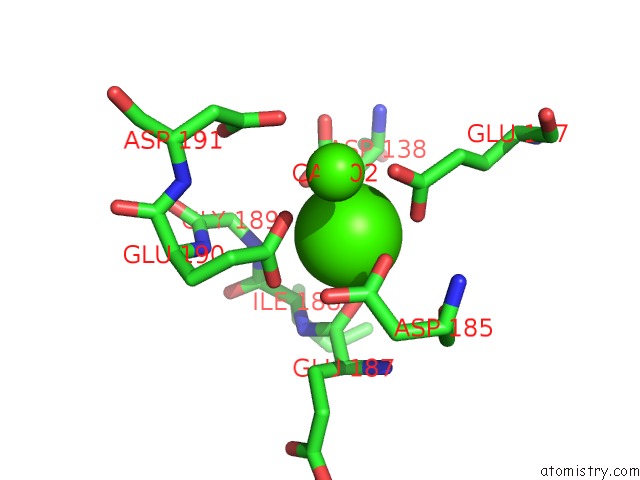
Mono view
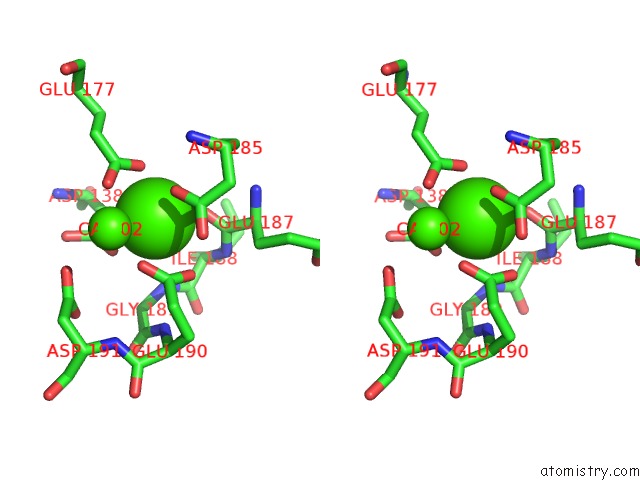
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of 3D Electron Diffraction Structure of Thermolysin From Bacillus Thermoproteolyticus within 5.0Å range:
|
Calcium binding site 2 out of 4 in 6zhj
Go back to
Calcium binding site 2 out
of 4 in the 3D Electron Diffraction Structure of Thermolysin From Bacillus Thermoproteolyticus
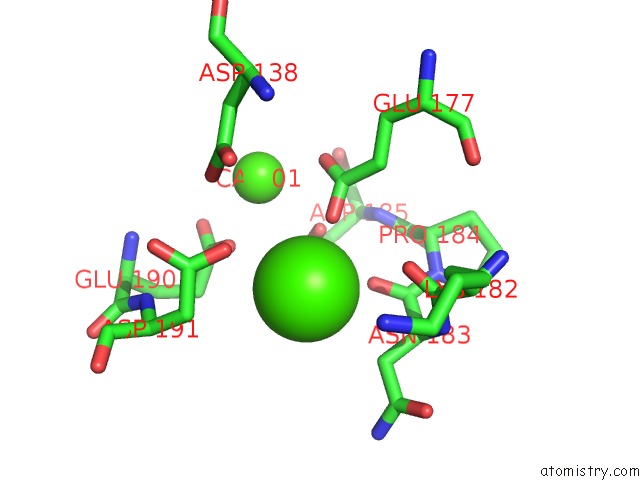
Mono view
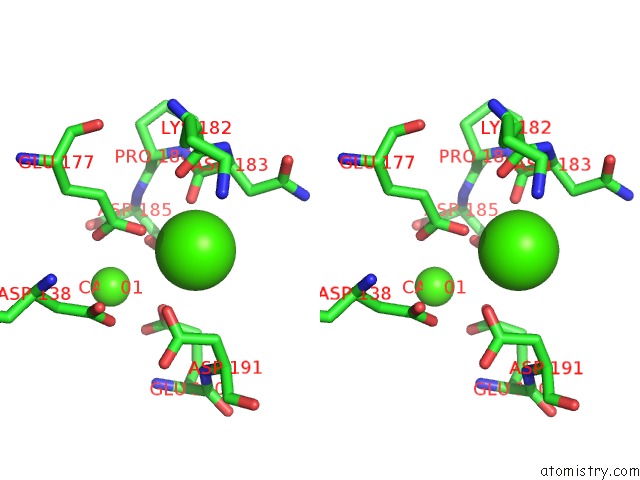
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of 3D Electron Diffraction Structure of Thermolysin From Bacillus Thermoproteolyticus within 5.0Å range:
|
Calcium binding site 3 out of 4 in 6zhj
Go back to
Calcium binding site 3 out
of 4 in the 3D Electron Diffraction Structure of Thermolysin From Bacillus Thermoproteolyticus
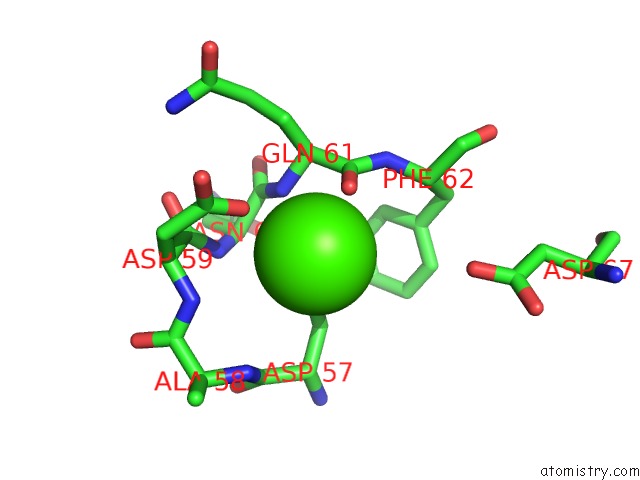
Mono view
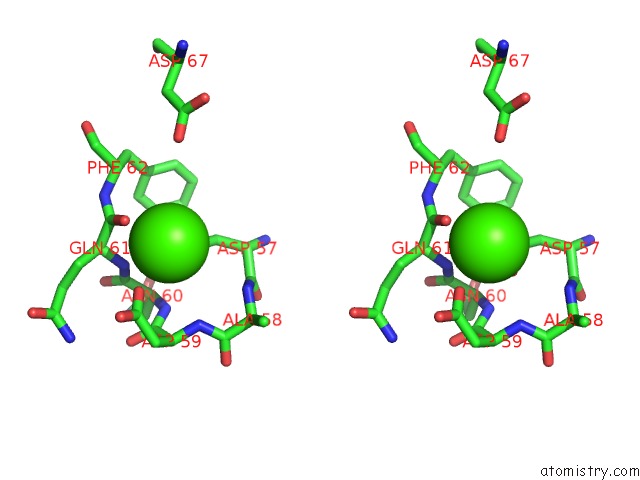
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of 3D Electron Diffraction Structure of Thermolysin From Bacillus Thermoproteolyticus within 5.0Å range:
|
Calcium binding site 4 out of 4 in 6zhj
Go back to
Calcium binding site 4 out
of 4 in the 3D Electron Diffraction Structure of Thermolysin From Bacillus Thermoproteolyticus
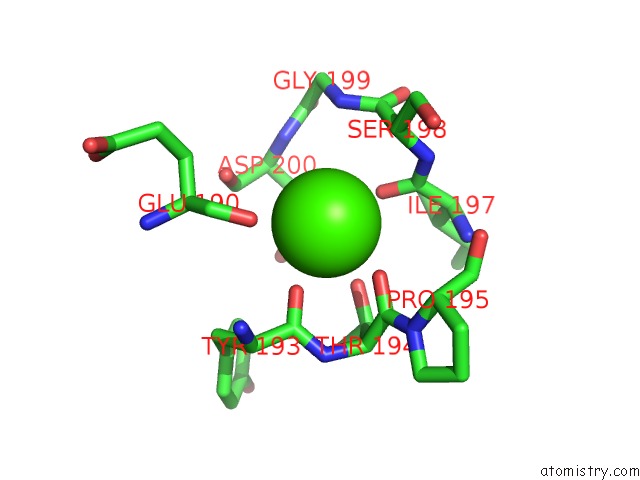
Mono view
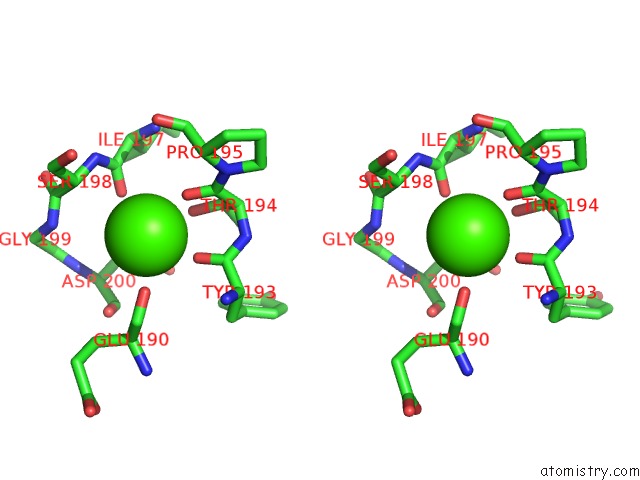
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of 3D Electron Diffraction Structure of Thermolysin From Bacillus Thermoproteolyticus within 5.0Å range:
|
Reference:
T.B.Blum,
D.Housset,
M.T.B.Clabbers,
E.Van Genderen,
M.Bacia-Verloop,
U.Zander,
A.A.Mccarthy,
G.Schoehn,
W.L.Ling,
J.P.Abrahams.
Statistically Correcting Dynamical Electron Scattering Improves the Refinement of Protein Nanocrystals, Including Charge Refinement of Coordinated Metals. Acta Crystallogr D Struct V. 77 75 2021BIOL.
ISSN: ISSN 2059-7983
PubMed: 33404527
DOI: 10.1107/S2059798320014540
Page generated: Thu Jul 18 22:40:23 2024
ISSN: ISSN 2059-7983
PubMed: 33404527
DOI: 10.1107/S2059798320014540
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1