Calcium »
PDB 7d52-7dl7 »
7dkw »
Calcium in PDB 7dkw: Crystal Structure of TXGH116 E441G Nucleophile Mutant From Thermoanaerobacterium Xylanolyticum with Autocondensation Products From Alpha-Fluoroglucoside.
Enzymatic activity of Crystal Structure of TXGH116 E441G Nucleophile Mutant From Thermoanaerobacterium Xylanolyticum with Autocondensation Products From Alpha-Fluoroglucoside.
All present enzymatic activity of Crystal Structure of TXGH116 E441G Nucleophile Mutant From Thermoanaerobacterium Xylanolyticum with Autocondensation Products From Alpha-Fluoroglucoside.:
3.2.1.21;
3.2.1.21;
Protein crystallography data
The structure of Crystal Structure of TXGH116 E441G Nucleophile Mutant From Thermoanaerobacterium Xylanolyticum with Autocondensation Products From Alpha-Fluoroglucoside., PDB code: 7dkw
was solved by
S.Pengthaisong,
J.R.Ketudat Cairns,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 35.00 / 1.78 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 179.214, 54.602, 164.55, 90, 90, 90 |
| R / Rfree (%) | 15.8 / 18.8 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of TXGH116 E441G Nucleophile Mutant From Thermoanaerobacterium Xylanolyticum with Autocondensation Products From Alpha-Fluoroglucoside.
(pdb code 7dkw). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Crystal Structure of TXGH116 E441G Nucleophile Mutant From Thermoanaerobacterium Xylanolyticum with Autocondensation Products From Alpha-Fluoroglucoside., PDB code: 7dkw:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Crystal Structure of TXGH116 E441G Nucleophile Mutant From Thermoanaerobacterium Xylanolyticum with Autocondensation Products From Alpha-Fluoroglucoside., PDB code: 7dkw:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 7dkw
Go back to
Calcium binding site 1 out
of 2 in the Crystal Structure of TXGH116 E441G Nucleophile Mutant From Thermoanaerobacterium Xylanolyticum with Autocondensation Products From Alpha-Fluoroglucoside.
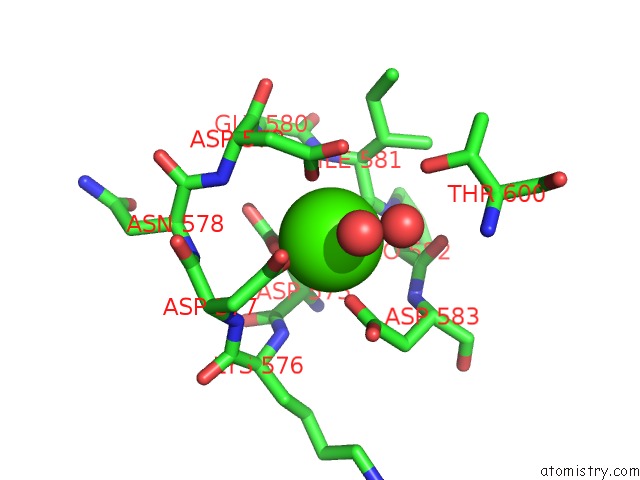
Mono view
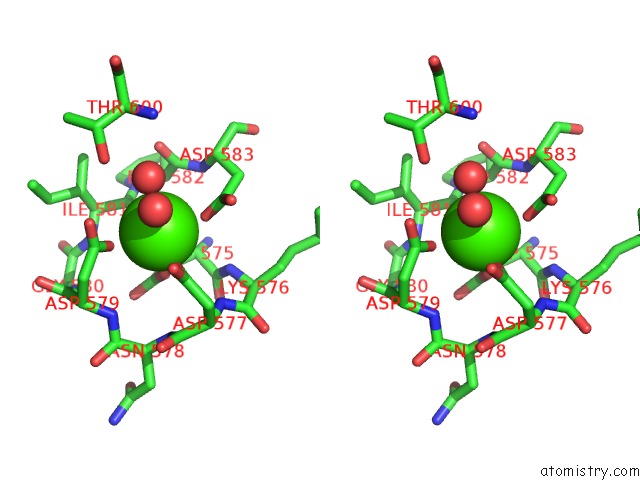
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of TXGH116 E441G Nucleophile Mutant From Thermoanaerobacterium Xylanolyticum with Autocondensation Products From Alpha-Fluoroglucoside. within 5.0Å range:
|
Calcium binding site 2 out of 2 in 7dkw
Go back to
Calcium binding site 2 out
of 2 in the Crystal Structure of TXGH116 E441G Nucleophile Mutant From Thermoanaerobacterium Xylanolyticum with Autocondensation Products From Alpha-Fluoroglucoside.
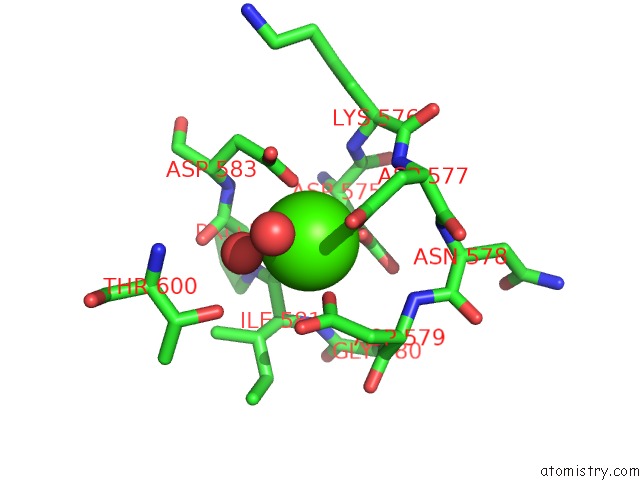
Mono view
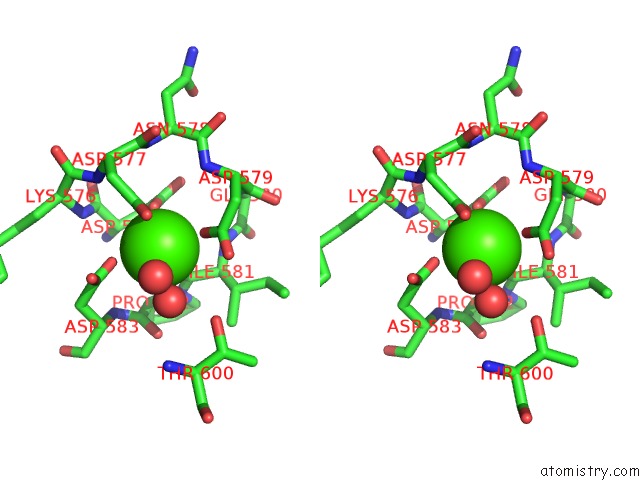
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Crystal Structure of TXGH116 E441G Nucleophile Mutant From Thermoanaerobacterium Xylanolyticum with Autocondensation Products From Alpha-Fluoroglucoside. within 5.0Å range:
|
Reference:
S.Pengthaisong,
Y.Hua,
J.R.Ketudat Cairns.
Structural Basis For Transglycosylation in Glycoside Hydrolase Family GH116 Glycosynthases. Arch.Biochem.Biophys. V. 706 08924 2021.
ISSN: ESSN 1096-0384
PubMed: 34019851
DOI: 10.1016/J.ABB.2021.108924
Page generated: Wed Jul 9 21:37:20 2025
ISSN: ESSN 1096-0384
PubMed: 34019851
DOI: 10.1016/J.ABB.2021.108924
Last articles
Cd in 4J0DCd in 4I7Z
Cd in 4HL1
Cd in 4HX7
Cd in 4H35
Cd in 4HKY
Cd in 4H3U
Cd in 4H3O
Cd in 4H44
Cd in 4H0L