Calcium »
PDB 7l74-7lrx »
7l8v »
Calcium in PDB 7l8v: uc(Nmr) Structure of Half-Calcified Calmodulin Mutant (CAMEF12) Bound to the Iq-Motif of CAV1.2
Calcium Binding Sites:
The binding sites of Calcium atom in the uc(Nmr) Structure of Half-Calcified Calmodulin Mutant (CAMEF12) Bound to the Iq-Motif of CAV1.2
(pdb code 7l8v). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the uc(Nmr) Structure of Half-Calcified Calmodulin Mutant (CAMEF12) Bound to the Iq-Motif of CAV1.2, PDB code: 7l8v:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the uc(Nmr) Structure of Half-Calcified Calmodulin Mutant (CAMEF12) Bound to the Iq-Motif of CAV1.2, PDB code: 7l8v:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 7l8v
Go back to
Calcium binding site 1 out
of 2 in the uc(Nmr) Structure of Half-Calcified Calmodulin Mutant (CAMEF12) Bound to the Iq-Motif of CAV1.2
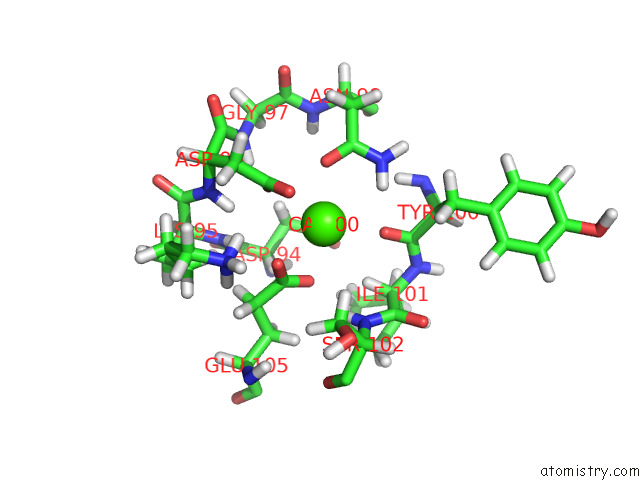
Mono view
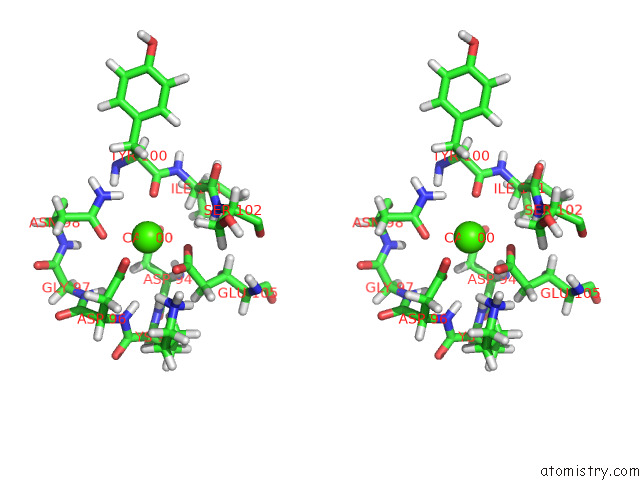
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of uc(Nmr) Structure of Half-Calcified Calmodulin Mutant (CAMEF12) Bound to the Iq-Motif of CAV1.2 within 5.0Å range:
|
Calcium binding site 2 out of 2 in 7l8v
Go back to
Calcium binding site 2 out
of 2 in the uc(Nmr) Structure of Half-Calcified Calmodulin Mutant (CAMEF12) Bound to the Iq-Motif of CAV1.2
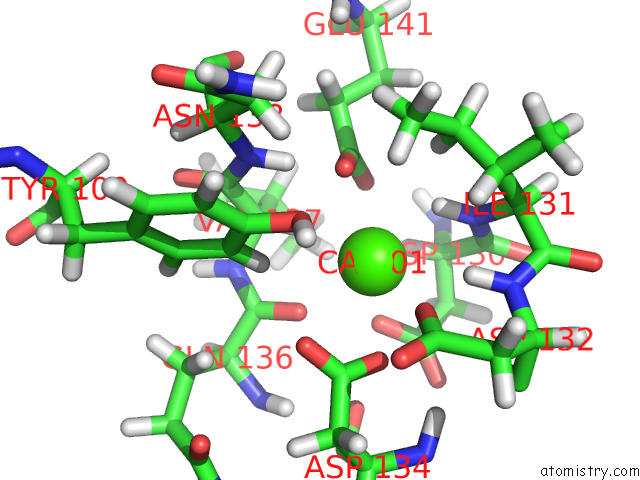
Mono view
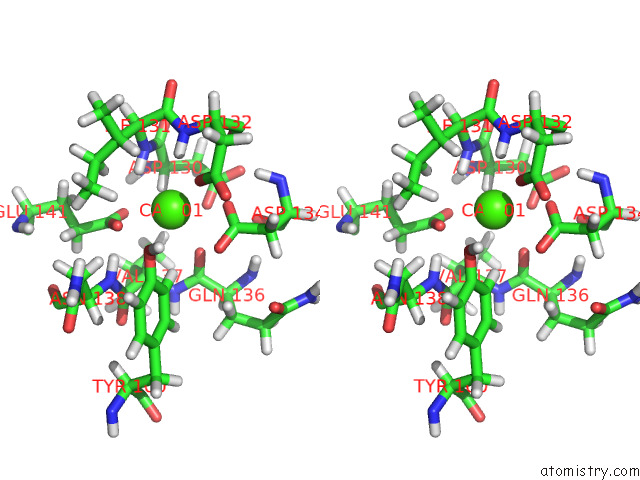
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of uc(Nmr) Structure of Half-Calcified Calmodulin Mutant (CAMEF12) Bound to the Iq-Motif of CAV1.2 within 5.0Å range:
|
Reference:
J.B.Ames,
J.B.Ames.
N/A N/A.
Page generated: Wed Jul 9 23:06:46 2025
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO