Calcium »
PDB 7skm-7tbg »
7t4s »
Calcium in PDB 7t4s: Cryoem Structure of the Hcmv Pentamer Gh/Gl/UL128/UL130/UL131A in Complex with NRP2 and Neutralizing Fabs 8I21 and 13H11
Calcium Binding Sites:
The binding sites of Calcium atom in the Cryoem Structure of the Hcmv Pentamer Gh/Gl/UL128/UL130/UL131A in Complex with NRP2 and Neutralizing Fabs 8I21 and 13H11
(pdb code 7t4s). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total only one binding site of Calcium was determined in the Cryoem Structure of the Hcmv Pentamer Gh/Gl/UL128/UL130/UL131A in Complex with NRP2 and Neutralizing Fabs 8I21 and 13H11, PDB code: 7t4s:
In total only one binding site of Calcium was determined in the Cryoem Structure of the Hcmv Pentamer Gh/Gl/UL128/UL130/UL131A in Complex with NRP2 and Neutralizing Fabs 8I21 and 13H11, PDB code: 7t4s:
Calcium binding site 1 out of 1 in 7t4s
Go back to
Calcium binding site 1 out
of 1 in the Cryoem Structure of the Hcmv Pentamer Gh/Gl/UL128/UL130/UL131A in Complex with NRP2 and Neutralizing Fabs 8I21 and 13H11
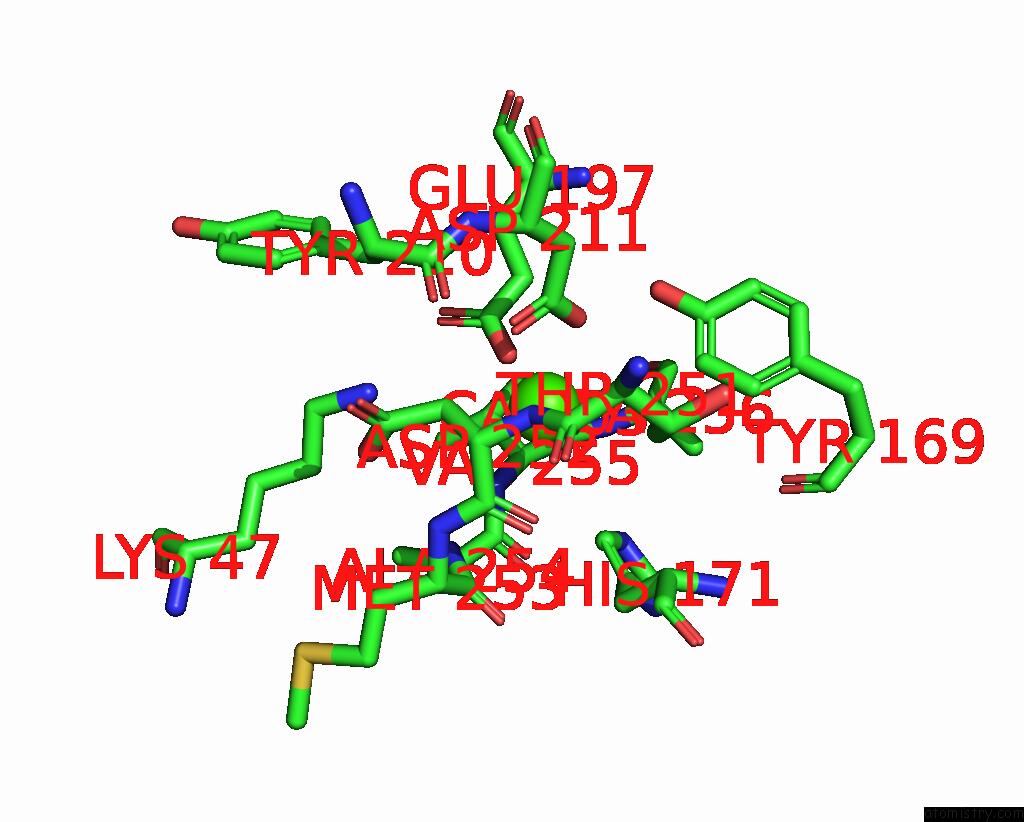
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Cryoem Structure of the Hcmv Pentamer Gh/Gl/UL128/UL130/UL131A in Complex with NRP2 and Neutralizing Fabs 8I21 and 13H11 within 5.0Å range:
|
Reference:
M.Kschonsak,
M.C.Johnson,
R.Schelling,
E.M.Green,
L.Rouge,
H.Ho,
N.Patel,
C.Kilic,
E.Kraft,
C.P.Arthur,
A.L.Rohou,
L.Comps-Agrar,
N.Martinez-Martin,
L.Perez,
J.Payandeh,
C.Ciferri.
Structural Basis For Hcmv Pentamer Receptor Recognition and Antibody Neutralization. Sci Adv V. 8 M2536 2022.
ISSN: ESSN 2375-2548
PubMed: 35275719
DOI: 10.1126/SCIADV.ABM2536
Page generated: Fri Jul 19 04:20:35 2024
ISSN: ESSN 2375-2548
PubMed: 35275719
DOI: 10.1126/SCIADV.ABM2536
Last articles
Zn in 9JYWZn in 9IR4
Zn in 9IR3
Zn in 9GMX
Zn in 9GMW
Zn in 9JEJ
Zn in 9ERF
Zn in 9ERE
Zn in 9EGV
Zn in 9EGW