Calcium »
PDB 7uww-7vmj »
7vg3 »
Calcium in PDB 7vg3: Cryo-Em Structure of Arabidopsis DCL3 in Complex with A 30-Bp Rna
Other elements in 7vg3:
The structure of Cryo-Em Structure of Arabidopsis DCL3 in Complex with A 30-Bp Rna also contains other interesting chemical elements:
| Zinc | (Zn) | 1 atom |
Calcium Binding Sites:
The binding sites of Calcium atom in the Cryo-Em Structure of Arabidopsis DCL3 in Complex with A 30-Bp Rna
(pdb code 7vg3). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Cryo-Em Structure of Arabidopsis DCL3 in Complex with A 30-Bp Rna, PDB code: 7vg3:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Cryo-Em Structure of Arabidopsis DCL3 in Complex with A 30-Bp Rna, PDB code: 7vg3:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 7vg3
Go back to
Calcium binding site 1 out
of 2 in the Cryo-Em Structure of Arabidopsis DCL3 in Complex with A 30-Bp Rna

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Cryo-Em Structure of Arabidopsis DCL3 in Complex with A 30-Bp Rna within 5.0Å range:
|
Calcium binding site 2 out of 2 in 7vg3
Go back to
Calcium binding site 2 out
of 2 in the Cryo-Em Structure of Arabidopsis DCL3 in Complex with A 30-Bp Rna

Mono view
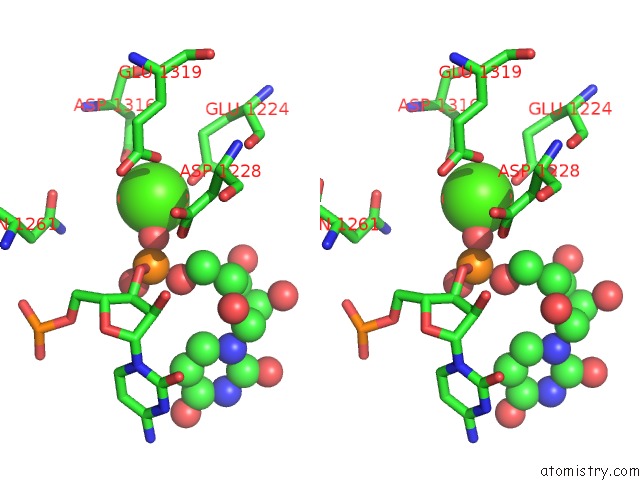
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Cryo-Em Structure of Arabidopsis DCL3 in Complex with A 30-Bp Rna within 5.0Å range:
|
Reference:
Q.Wang,
Y.Xue,
L.Zhang,
Z.Zhong,
S.Feng,
C.Wang,
L.Xiao,
Z.Yang,
C.J.Harris,
Z.Wu,
J.Zhai,
M.Yang,
S.Li,
S.E.Jacobsen,
J.Du.
Mechanism of Sirna Production By A Plant Dicer-Rna Complex in Dicing-Competent Conformation. Science L4546 2021.
ISSN: ESSN 1095-9203
PubMed: 34648373
DOI: 10.1126/SCIENCE.ABL4546
Page generated: Fri Jul 19 05:19:24 2024
ISSN: ESSN 1095-9203
PubMed: 34648373
DOI: 10.1126/SCIENCE.ABL4546
Last articles
Zn in 9JYWZn in 9IR4
Zn in 9IR3
Zn in 9GMX
Zn in 9GMW
Zn in 9JEJ
Zn in 9ERF
Zn in 9ERE
Zn in 9EGV
Zn in 9EGW