Calcium »
PDB 7wxx-7xvv »
7x6c »
Calcium in PDB 7x6c: Cryo-Em Structure of the Human TRPC5 Ion Channel in Lipid Nanodiscs, CLASS1
Other elements in 7x6c:
The structure of Cryo-Em Structure of the Human TRPC5 Ion Channel in Lipid Nanodiscs, CLASS1 also contains other interesting chemical elements:
| Zinc | (Zn) | 4 atoms |
Calcium Binding Sites:
The binding sites of Calcium atom in the Cryo-Em Structure of the Human TRPC5 Ion Channel in Lipid Nanodiscs, CLASS1
(pdb code 7x6c). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 4 binding sites of Calcium where determined in the Cryo-Em Structure of the Human TRPC5 Ion Channel in Lipid Nanodiscs, CLASS1, PDB code: 7x6c:
Jump to Calcium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Calcium where determined in the Cryo-Em Structure of the Human TRPC5 Ion Channel in Lipid Nanodiscs, CLASS1, PDB code: 7x6c:
Jump to Calcium binding site number: 1; 2; 3; 4;
Calcium binding site 1 out of 4 in 7x6c
Go back to
Calcium binding site 1 out
of 4 in the Cryo-Em Structure of the Human TRPC5 Ion Channel in Lipid Nanodiscs, CLASS1
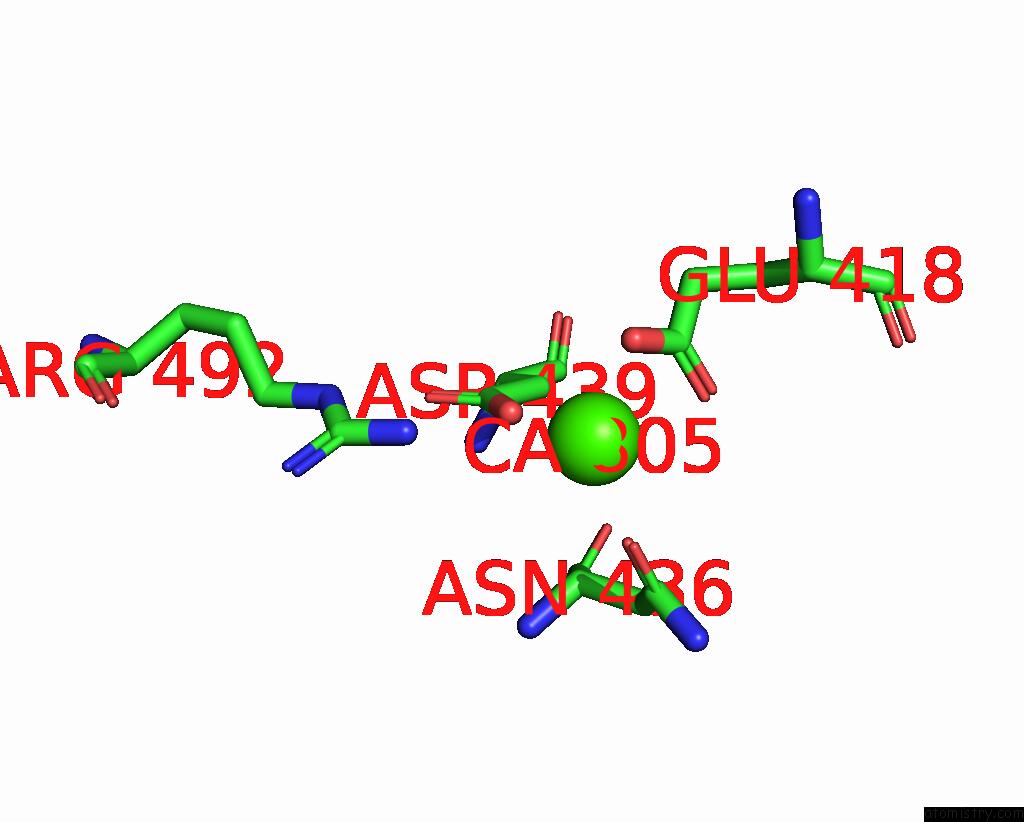
Mono view
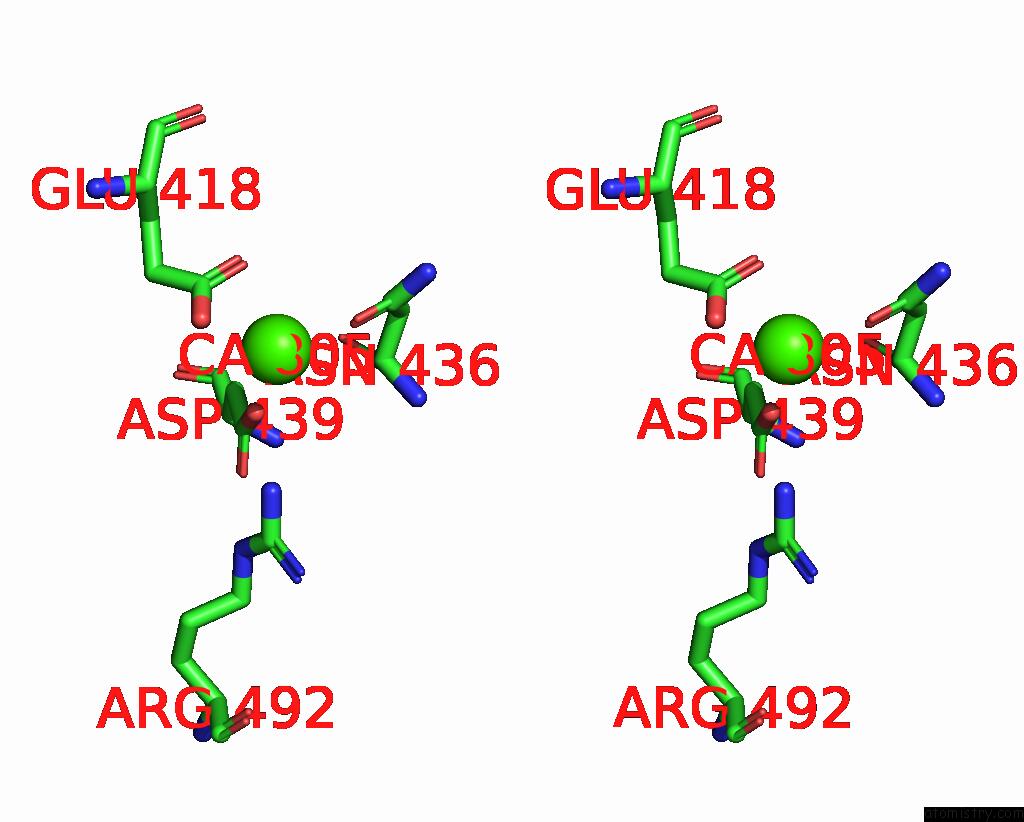
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Cryo-Em Structure of the Human TRPC5 Ion Channel in Lipid Nanodiscs, CLASS1 within 5.0Å range:
|
Calcium binding site 2 out of 4 in 7x6c
Go back to
Calcium binding site 2 out
of 4 in the Cryo-Em Structure of the Human TRPC5 Ion Channel in Lipid Nanodiscs, CLASS1
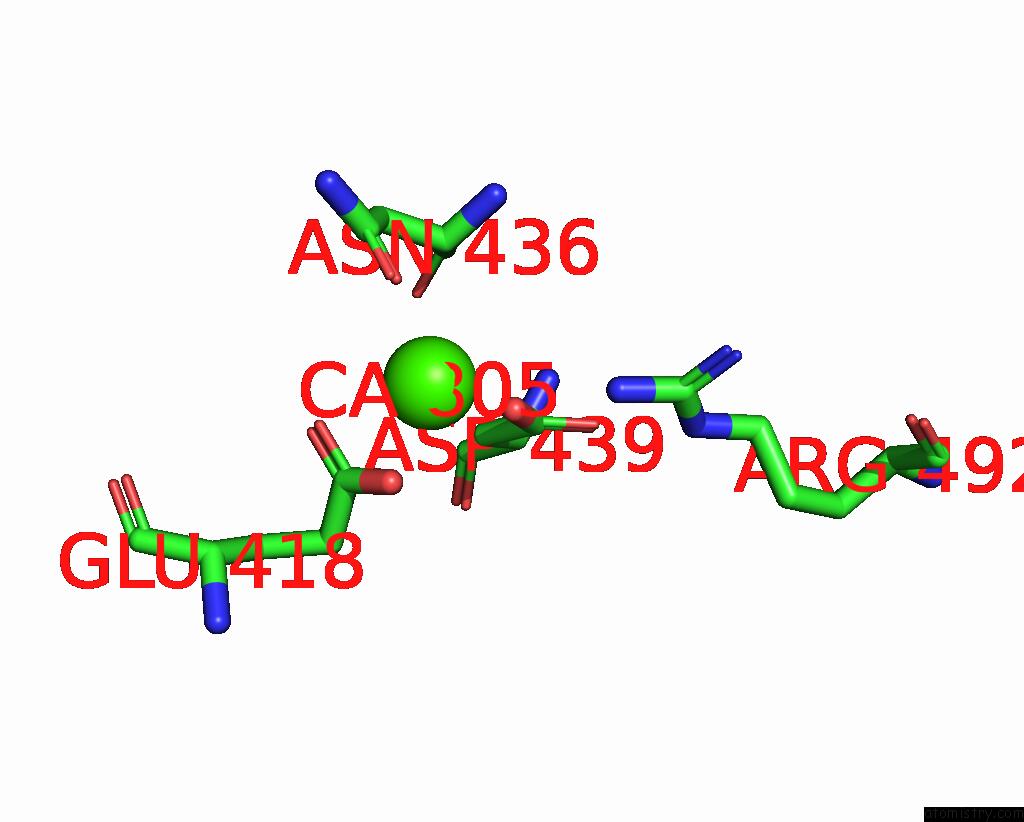
Mono view
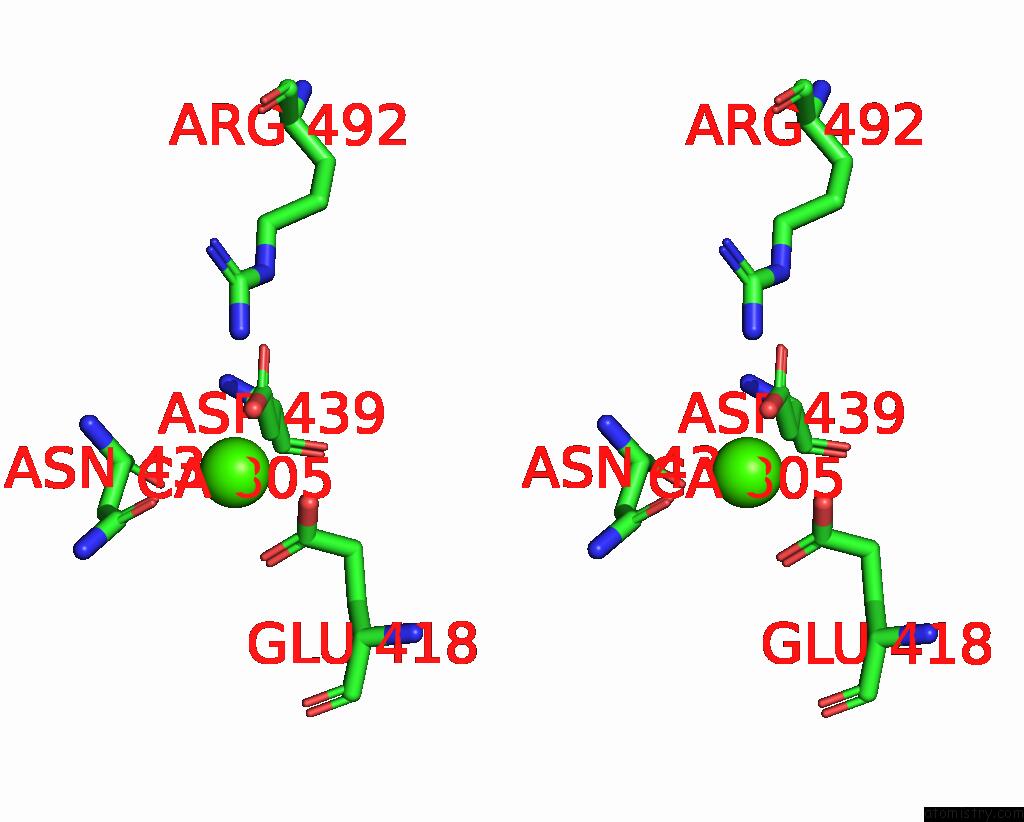
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Cryo-Em Structure of the Human TRPC5 Ion Channel in Lipid Nanodiscs, CLASS1 within 5.0Å range:
|
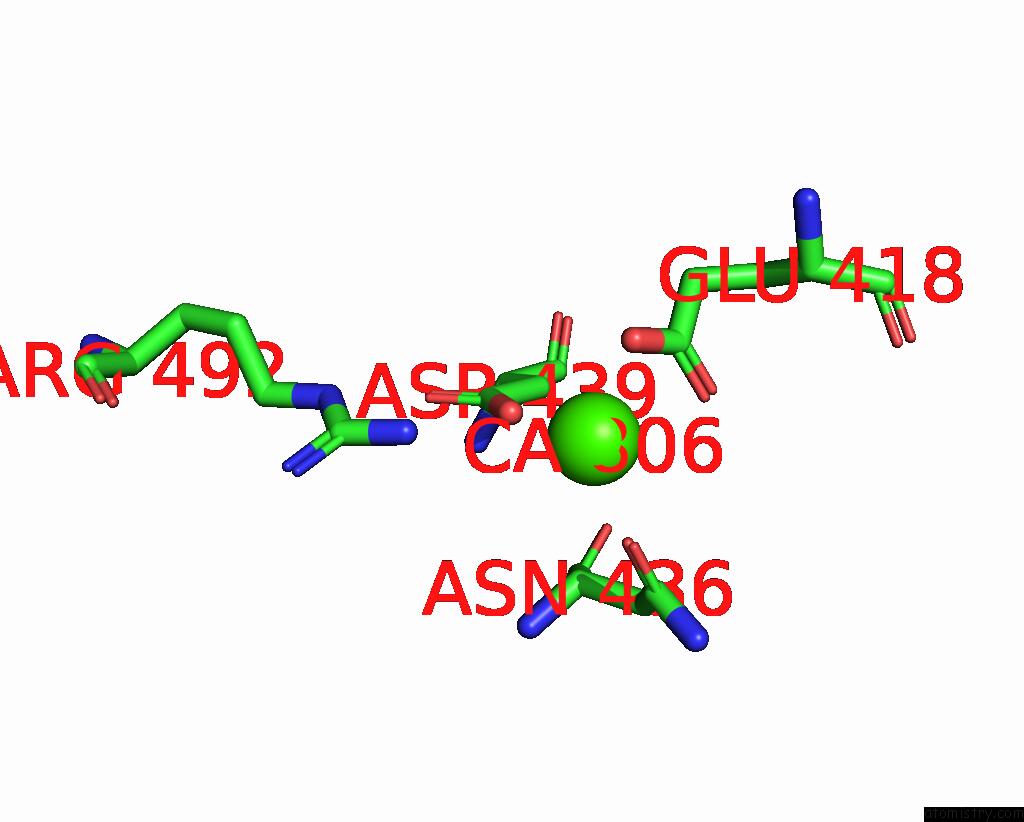
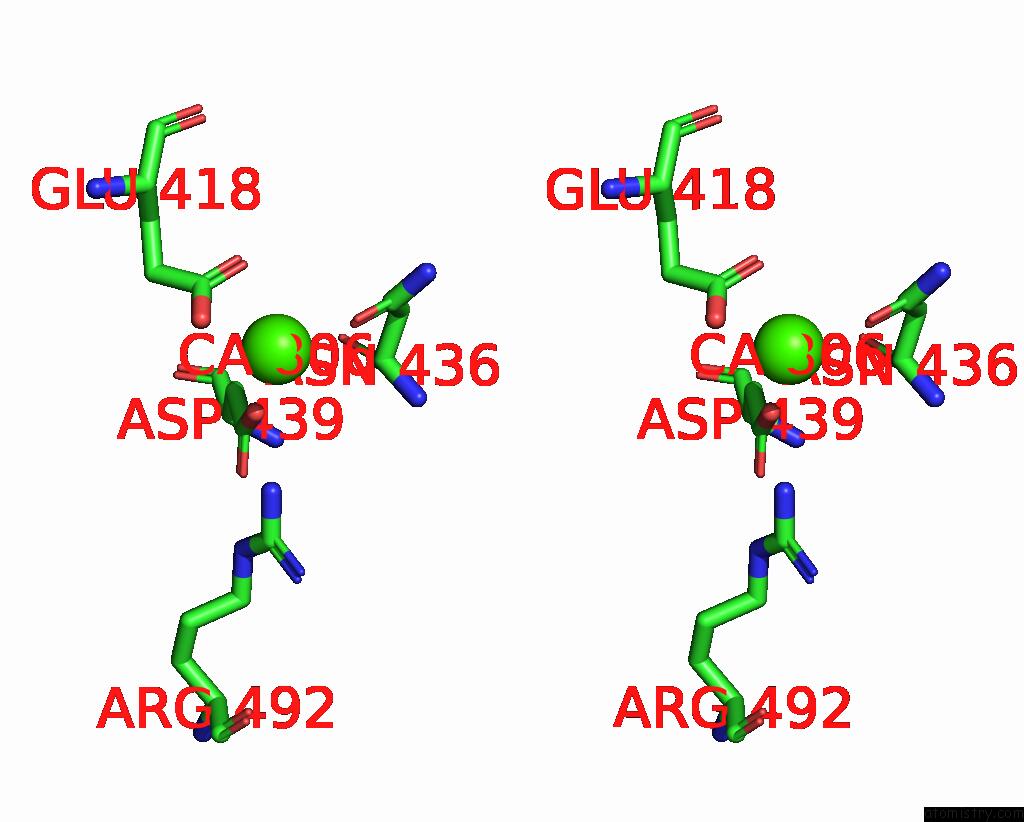
Calcium binding site 3 out of 4 in 7x6c
Go back to
Calcium binding site 3 out
of 4 in the Cryo-Em Structure of the Human TRPC5 Ion Channel in Lipid Nanodiscs, CLASS1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Cryo-Em Structure of the Human TRPC5 Ion Channel in Lipid Nanodiscs, CLASS1 within 5.0Å range:
|
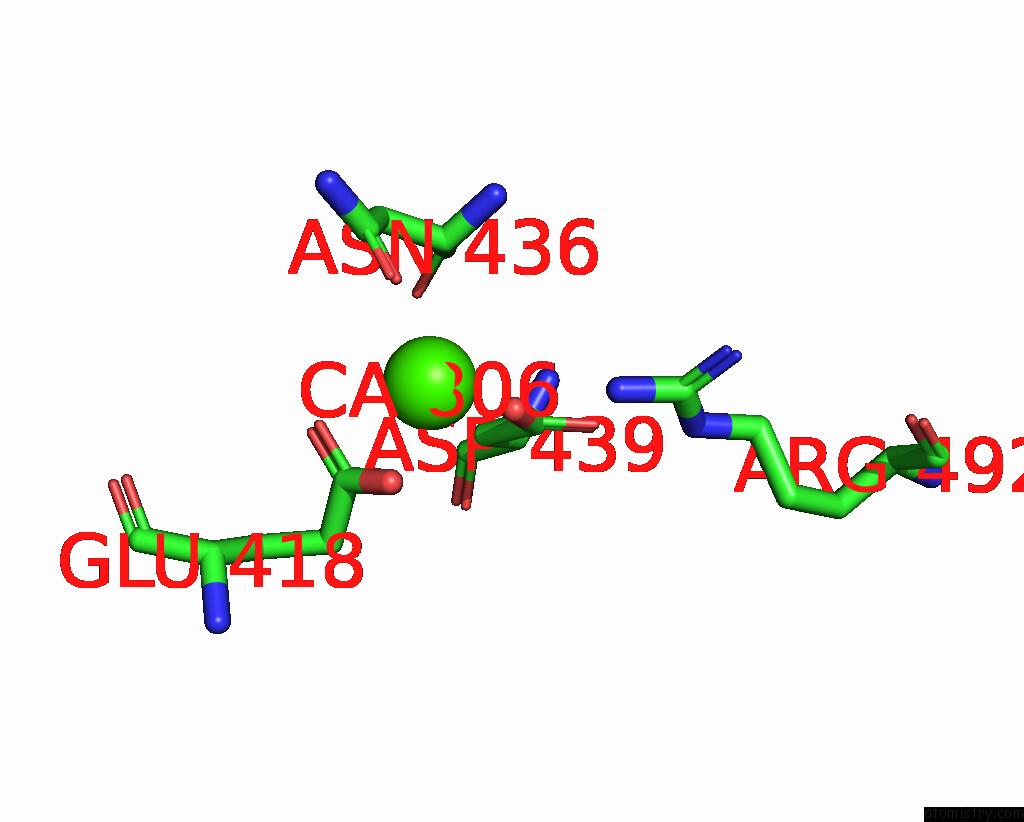
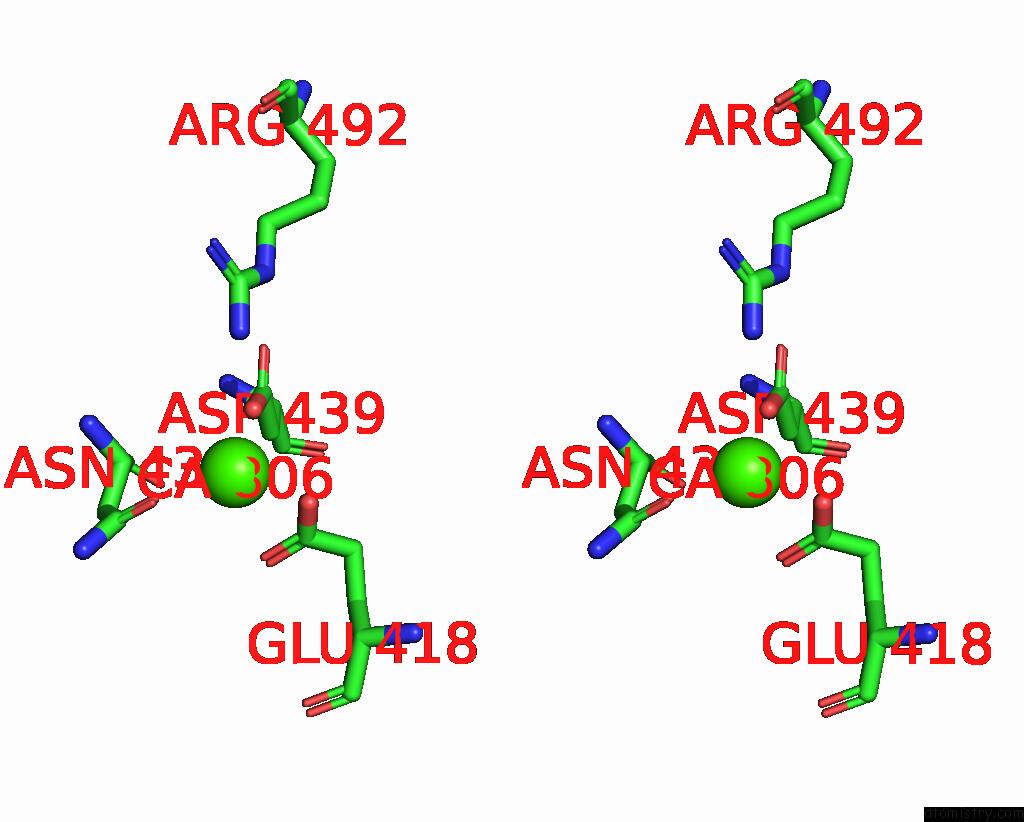
Calcium binding site 4 out of 4 in 7x6c
Go back to
Calcium binding site 4 out
of 4 in the Cryo-Em Structure of the Human TRPC5 Ion Channel in Lipid Nanodiscs, CLASS1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of Cryo-Em Structure of the Human TRPC5 Ion Channel in Lipid Nanodiscs, CLASS1 within 5.0Å range:
|
Reference:
J.Won,
J.Kim,
H.Jeong,
J.Kim,
S.Feng,
B.Jeong,
M.Kwak,
J.Ko,
W.Im,
I.So,
H.H.Lee.
Molecular Architecture of the G Alpha I -Bound TRPC5 Ion Channel. Nat Commun V. 14 2550 2023.
ISSN: ESSN 2041-1723
PubMed: 37137991
DOI: 10.1038/S41467-023-38281-3
Page generated: Thu Jul 10 02:25:39 2025
ISSN: ESSN 2041-1723
PubMed: 37137991
DOI: 10.1038/S41467-023-38281-3
Last articles
Cl in 5I1FCl in 5I0D
Cl in 5I13
Cl in 5I1C
Cl in 5I0W
Cl in 5I0V
Cl in 5HZX
Cl in 5I0R
Cl in 5I02
Cl in 5I00