Calcium »
PDB 8ax0-8bad »
8b79 »
Calcium in PDB 8b79: The Crystal Structure of M644G Variant of Dna Pol Epsilon Containing Utp in the Polymerase Active Site
Enzymatic activity of The Crystal Structure of M644G Variant of Dna Pol Epsilon Containing Utp in the Polymerase Active Site
All present enzymatic activity of The Crystal Structure of M644G Variant of Dna Pol Epsilon Containing Utp in the Polymerase Active Site:
2.7.7.7;
2.7.7.7;
Protein crystallography data
The structure of The Crystal Structure of M644G Variant of Dna Pol Epsilon Containing Utp in the Polymerase Active Site, PDB code: 8b79
was solved by
V.Parkash,
E.Johansson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 43.54 / 2.65 |
| Space group | P 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 154.4, 70.7, 159.4, 90, 112.8, 90 |
| R / Rfree (%) | 21.5 / 24.8 |
Calcium Binding Sites:
The binding sites of Calcium atom in the The Crystal Structure of M644G Variant of Dna Pol Epsilon Containing Utp in the Polymerase Active Site
(pdb code 8b79). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 6 binding sites of Calcium where determined in the The Crystal Structure of M644G Variant of Dna Pol Epsilon Containing Utp in the Polymerase Active Site, PDB code: 8b79:
Jump to Calcium binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Calcium where determined in the The Crystal Structure of M644G Variant of Dna Pol Epsilon Containing Utp in the Polymerase Active Site, PDB code: 8b79:
Jump to Calcium binding site number: 1; 2; 3; 4; 5; 6;
Calcium binding site 1 out of 6 in 8b79
Go back to
Calcium binding site 1 out
of 6 in the The Crystal Structure of M644G Variant of Dna Pol Epsilon Containing Utp in the Polymerase Active Site

Mono view
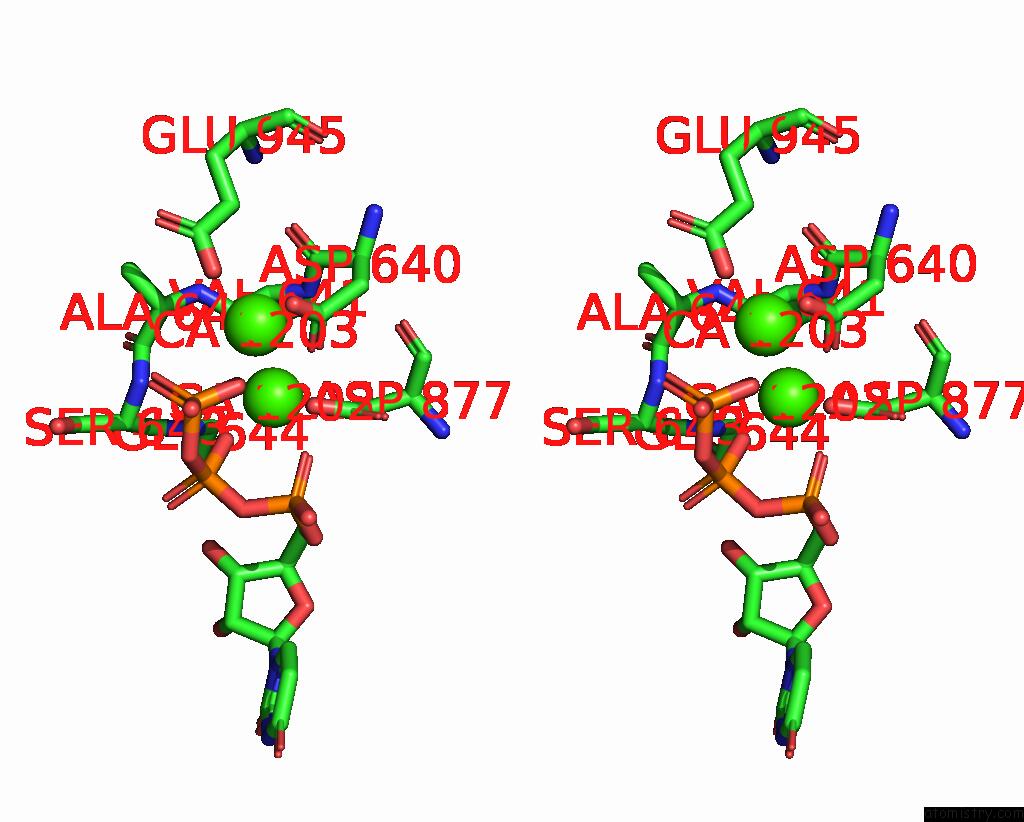
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of The Crystal Structure of M644G Variant of Dna Pol Epsilon Containing Utp in the Polymerase Active Site within 5.0Å range:
|
Calcium binding site 2 out of 6 in 8b79
Go back to
Calcium binding site 2 out
of 6 in the The Crystal Structure of M644G Variant of Dna Pol Epsilon Containing Utp in the Polymerase Active Site
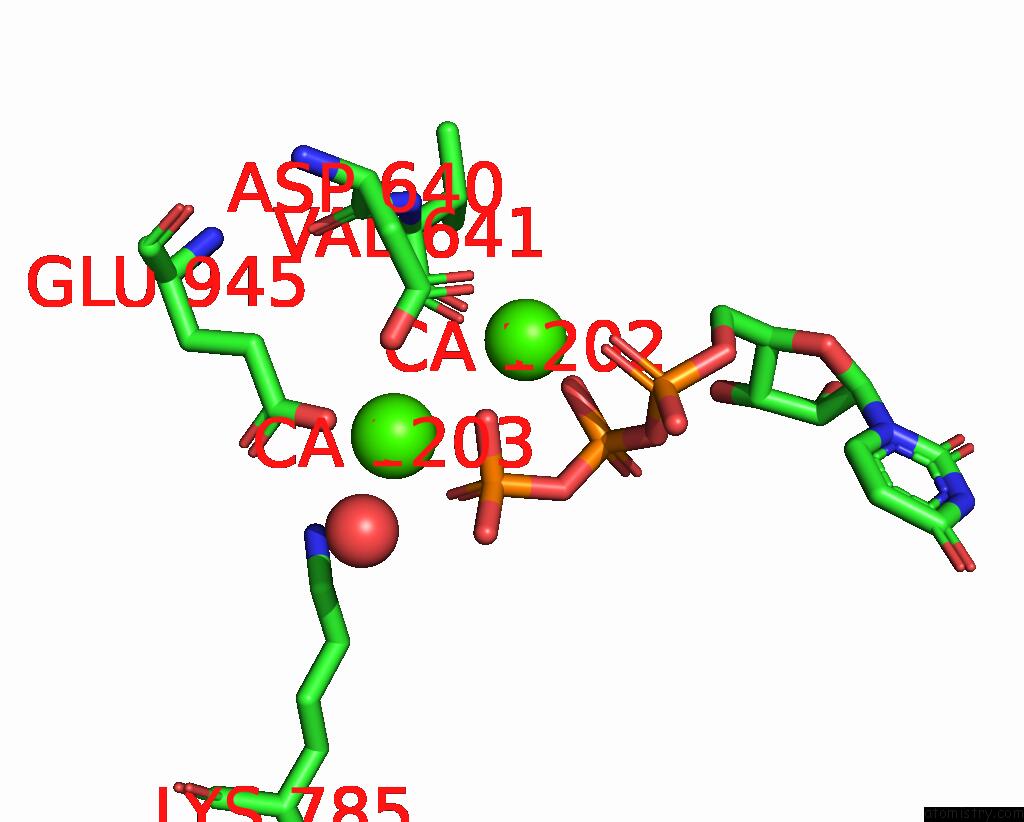
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of The Crystal Structure of M644G Variant of Dna Pol Epsilon Containing Utp in the Polymerase Active Site within 5.0Å range:
|
Calcium binding site 3 out of 6 in 8b79
Go back to
Calcium binding site 3 out
of 6 in the The Crystal Structure of M644G Variant of Dna Pol Epsilon Containing Utp in the Polymerase Active Site

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of The Crystal Structure of M644G Variant of Dna Pol Epsilon Containing Utp in the Polymerase Active Site within 5.0Å range:
|
Calcium binding site 4 out of 6 in 8b79
Go back to
Calcium binding site 4 out
of 6 in the The Crystal Structure of M644G Variant of Dna Pol Epsilon Containing Utp in the Polymerase Active Site

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of The Crystal Structure of M644G Variant of Dna Pol Epsilon Containing Utp in the Polymerase Active Site within 5.0Å range:
|
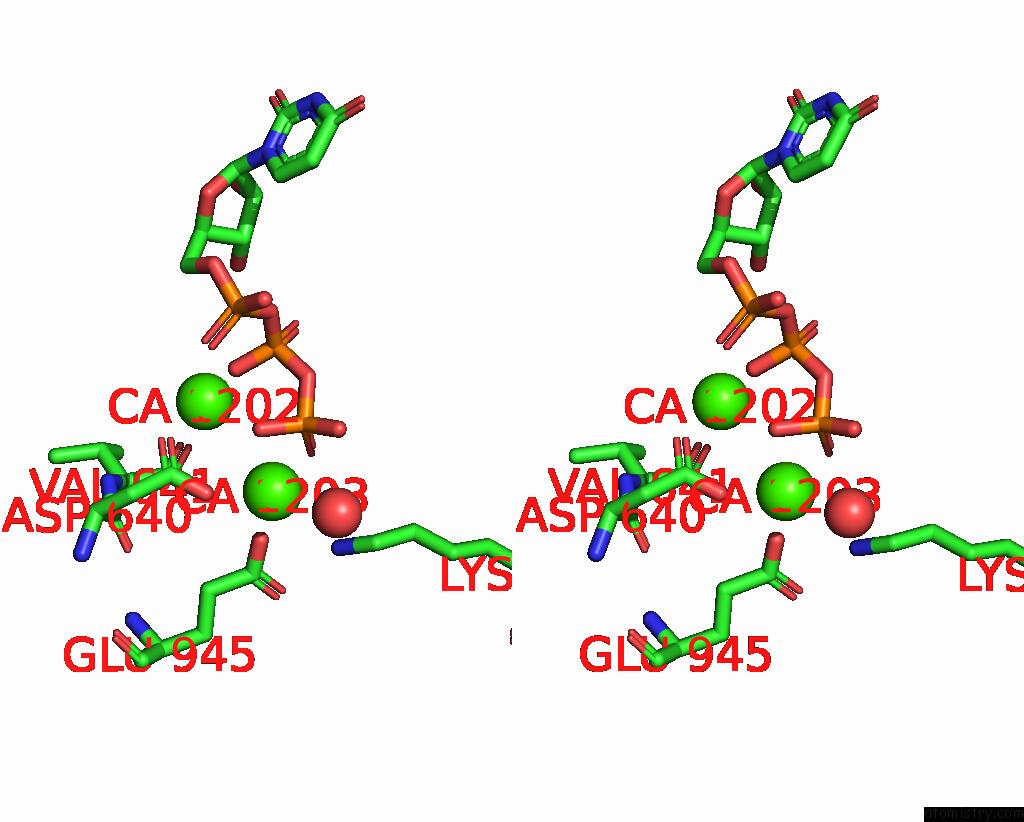
Calcium binding site 5 out of 6 in 8b79
Go back to
Calcium binding site 5 out
of 6 in the The Crystal Structure of M644G Variant of Dna Pol Epsilon Containing Utp in the Polymerase Active Site
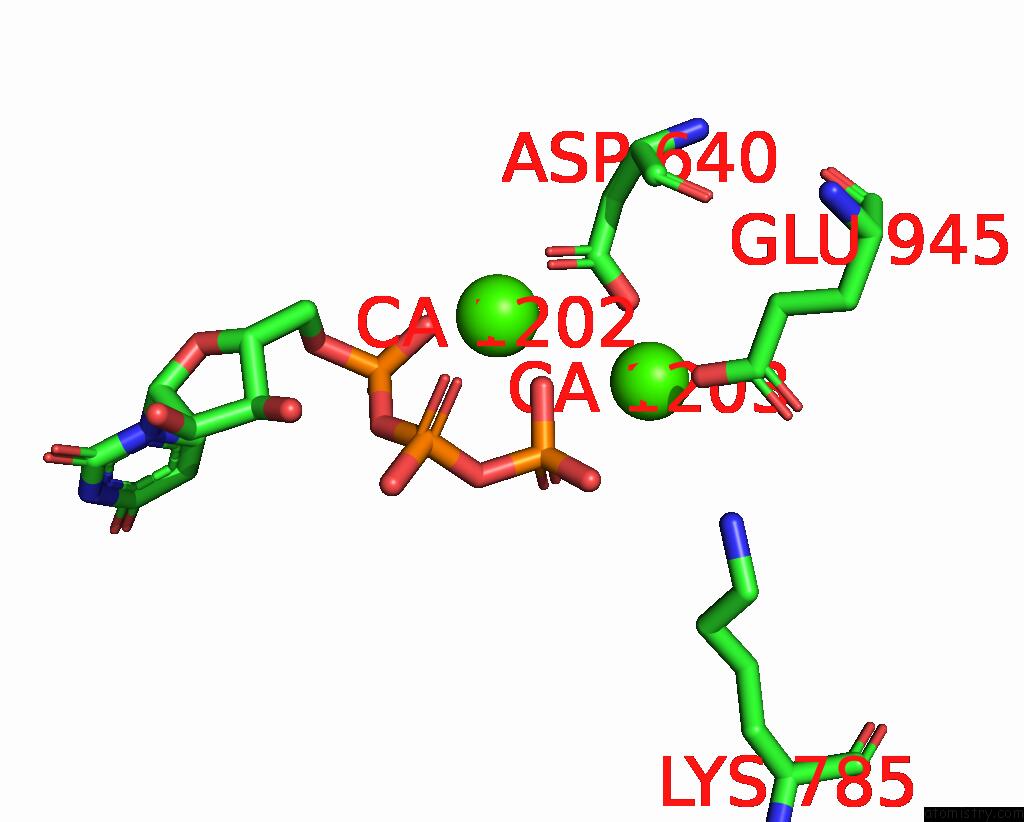
Mono view
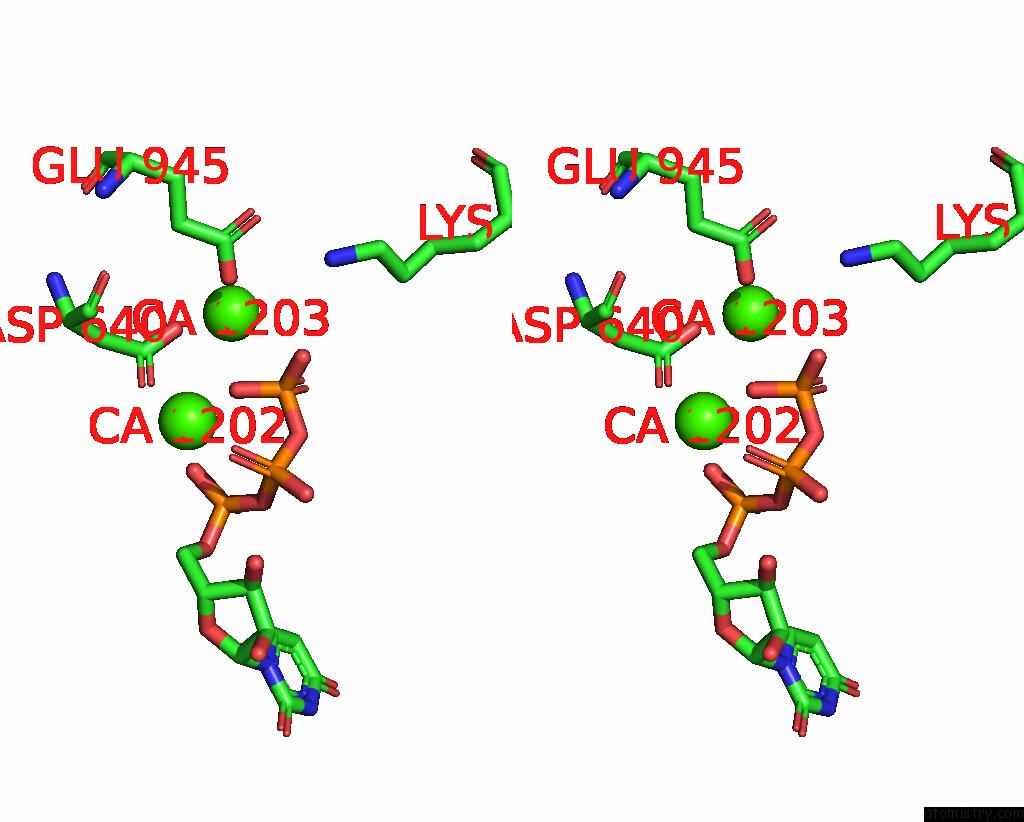
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 5 of The Crystal Structure of M644G Variant of Dna Pol Epsilon Containing Utp in the Polymerase Active Site within 5.0Å range:
|
Calcium binding site 6 out of 6 in 8b79
Go back to
Calcium binding site 6 out
of 6 in the The Crystal Structure of M644G Variant of Dna Pol Epsilon Containing Utp in the Polymerase Active Site
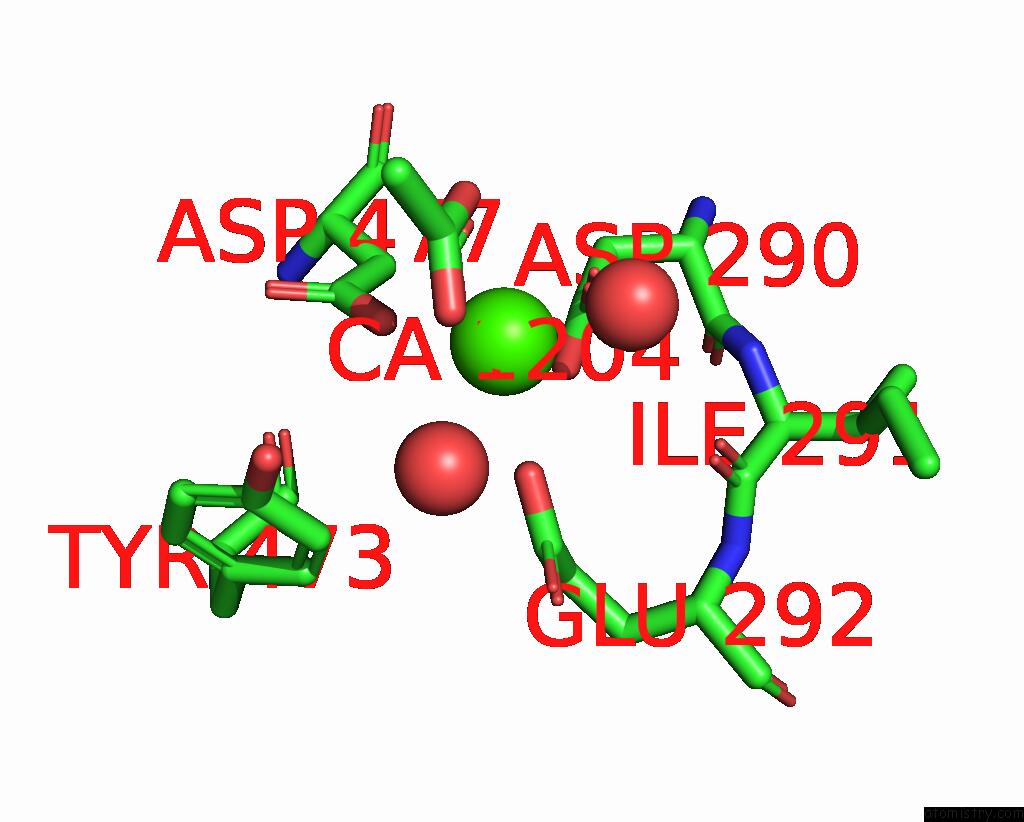
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 6 of The Crystal Structure of M644G Variant of Dna Pol Epsilon Containing Utp in the Polymerase Active Site within 5.0Å range:
|
Reference:
V.Parkash,
Y.Kulkarni,
G.O.Bylund,
P.Osterman,
A.Barrozo,
S.L.Kamerlin,
E.Johansson.
A Sensor Complements the Steric Gate When Dna Polymerase Epsilon Discriminates Ribonucleotides. To Be Published.
Page generated: Thu Jul 10 03:21:34 2025
Last articles
Cd in 4J0DCd in 4I7Z
Cd in 4HL1
Cd in 4HX7
Cd in 4H35
Cd in 4HKY
Cd in 4H3U
Cd in 4H3O
Cd in 4H44
Cd in 4H0L