Calcium »
PDB 8ax0-8bad »
8b8j »
Calcium in PDB 8b8j: Cryo-Em Structure of CA2+-Bound MTMEM16F F518H Mutant in Digitonin
Calcium Binding Sites:
The binding sites of Calcium atom in the Cryo-Em Structure of CA2+-Bound MTMEM16F F518H Mutant in Digitonin
(pdb code 8b8j). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 6 binding sites of Calcium where determined in the Cryo-Em Structure of CA2+-Bound MTMEM16F F518H Mutant in Digitonin, PDB code: 8b8j:
Jump to Calcium binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Calcium where determined in the Cryo-Em Structure of CA2+-Bound MTMEM16F F518H Mutant in Digitonin, PDB code: 8b8j:
Jump to Calcium binding site number: 1; 2; 3; 4; 5; 6;
Calcium binding site 1 out of 6 in 8b8j
Go back to
Calcium binding site 1 out
of 6 in the Cryo-Em Structure of CA2+-Bound MTMEM16F F518H Mutant in Digitonin
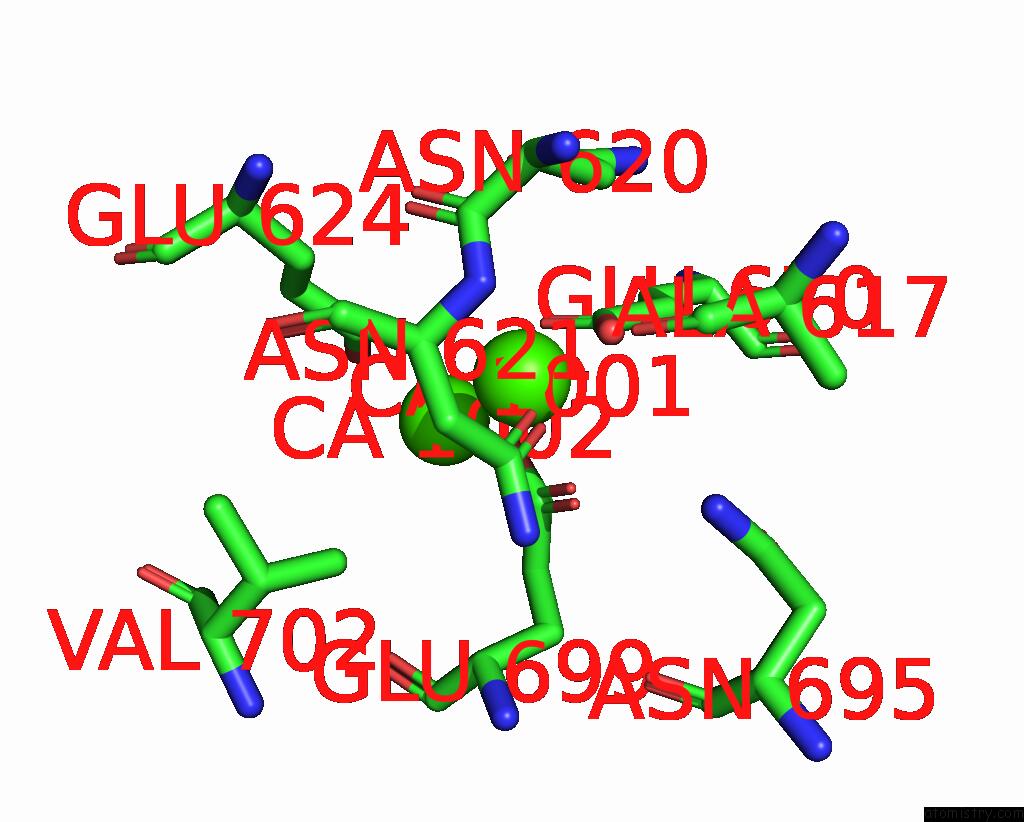
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Cryo-Em Structure of CA2+-Bound MTMEM16F F518H Mutant in Digitonin within 5.0Å range:
|
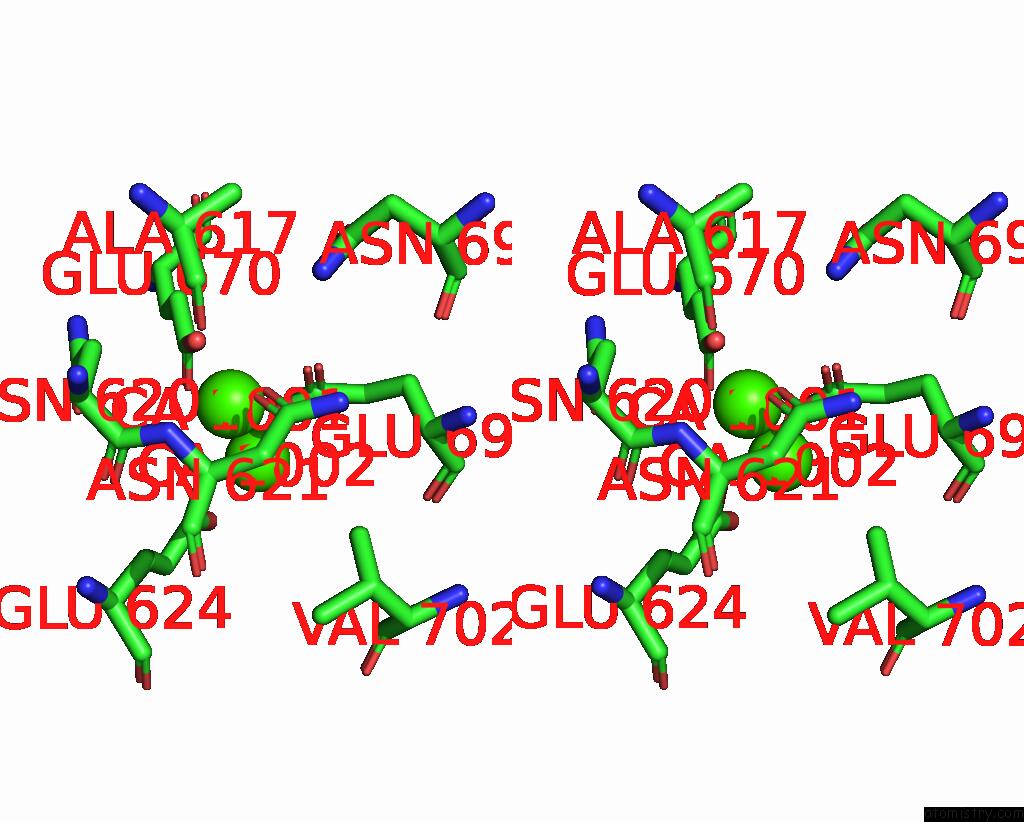
Calcium binding site 2 out of 6 in 8b8j
Go back to
Calcium binding site 2 out
of 6 in the Cryo-Em Structure of CA2+-Bound MTMEM16F F518H Mutant in Digitonin

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Cryo-Em Structure of CA2+-Bound MTMEM16F F518H Mutant in Digitonin within 5.0Å range:
|
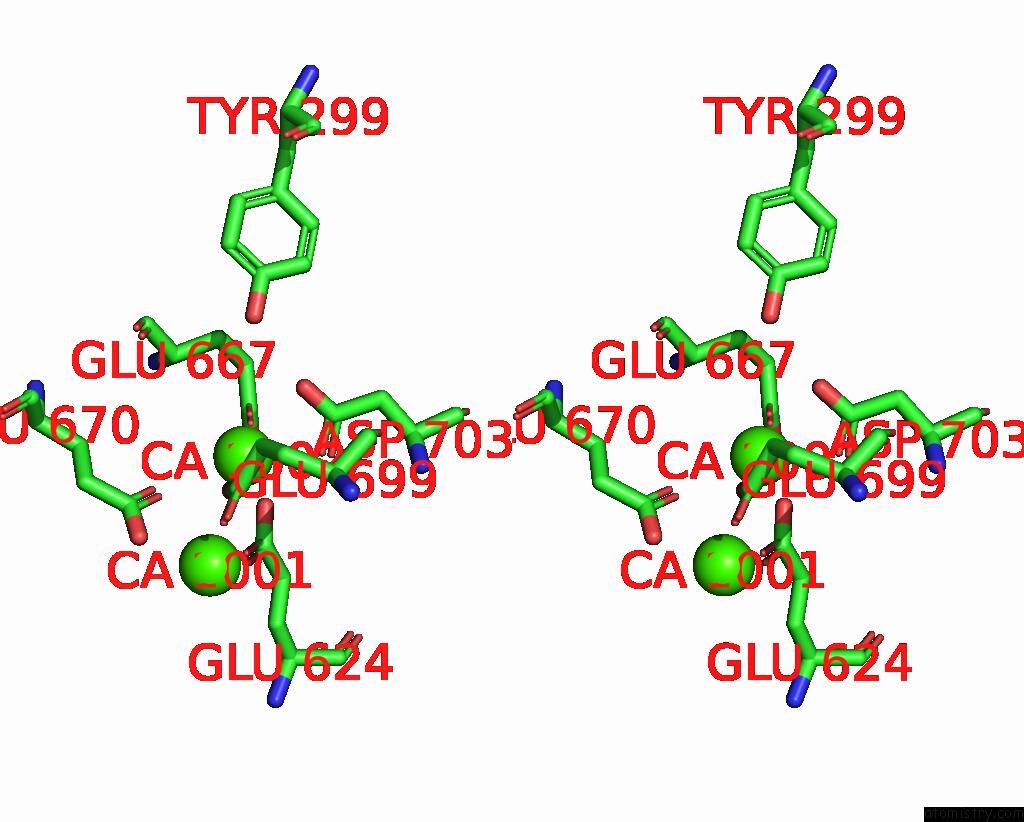
Calcium binding site 3 out of 6 in 8b8j
Go back to
Calcium binding site 3 out
of 6 in the Cryo-Em Structure of CA2+-Bound MTMEM16F F518H Mutant in Digitonin
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Cryo-Em Structure of CA2+-Bound MTMEM16F F518H Mutant in Digitonin within 5.0Å range:
|
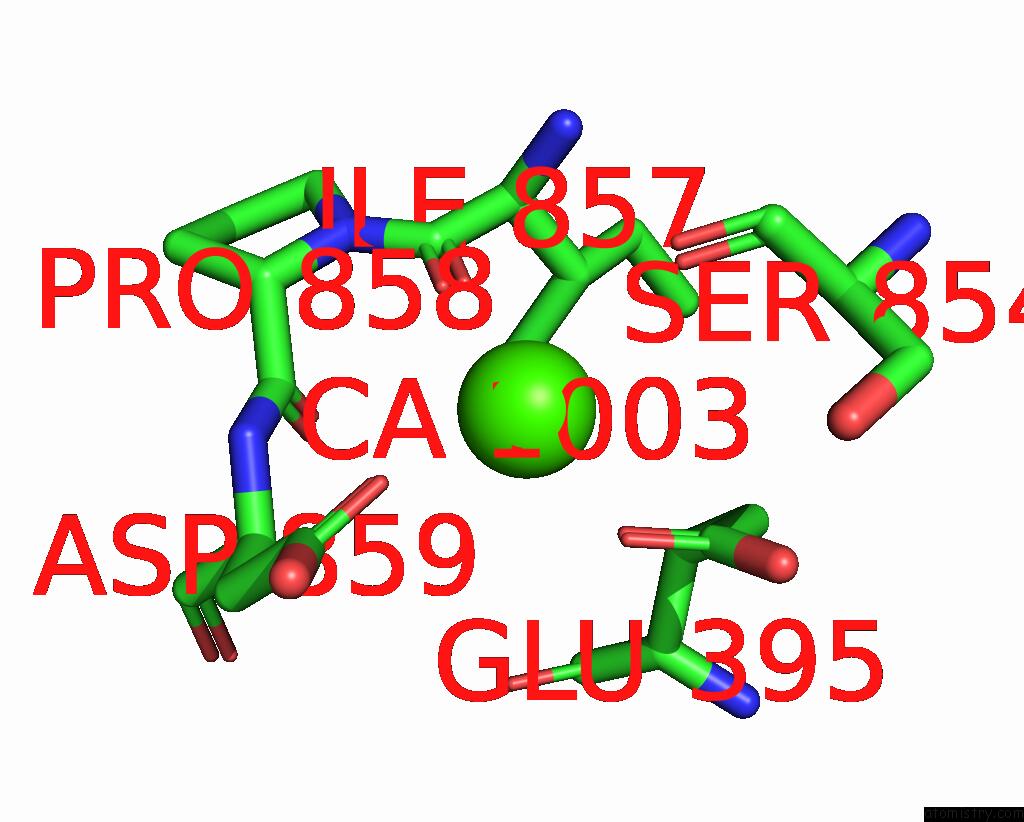
Calcium binding site 4 out of 6 in 8b8j
Go back to
Calcium binding site 4 out
of 6 in the Cryo-Em Structure of CA2+-Bound MTMEM16F F518H Mutant in Digitonin
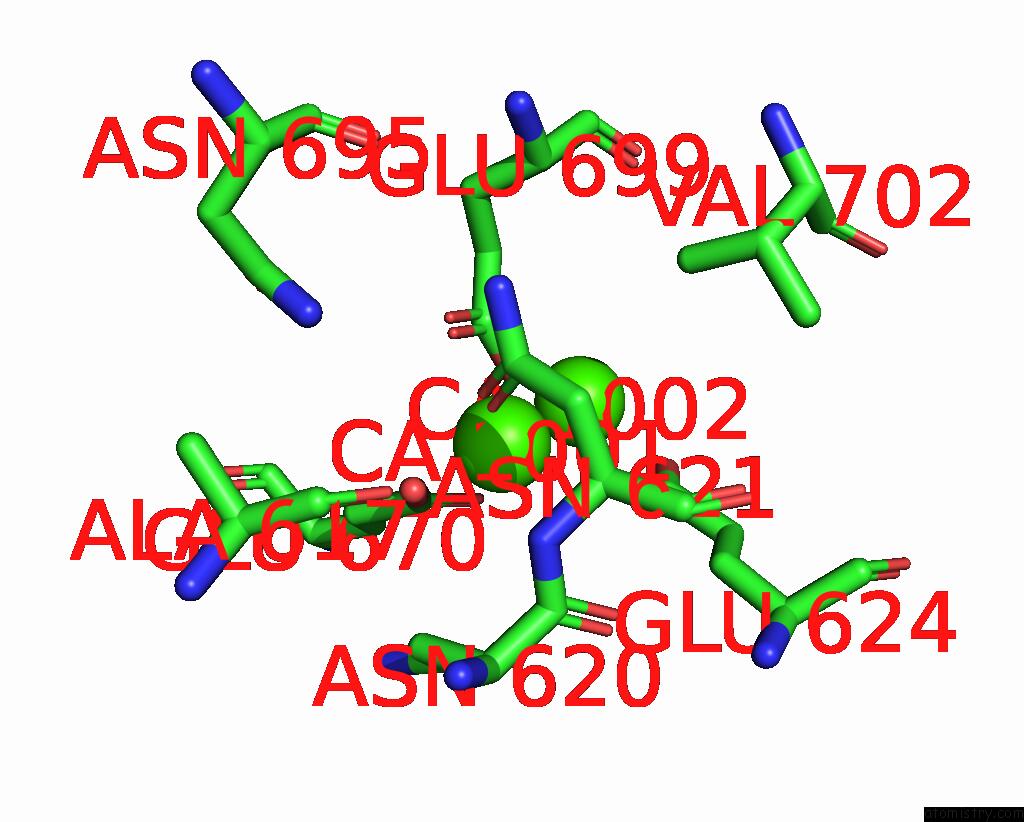
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of Cryo-Em Structure of CA2+-Bound MTMEM16F F518H Mutant in Digitonin within 5.0Å range:
|
Calcium binding site 5 out of 6 in 8b8j
Go back to
Calcium binding site 5 out
of 6 in the Cryo-Em Structure of CA2+-Bound MTMEM16F F518H Mutant in Digitonin

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 5 of Cryo-Em Structure of CA2+-Bound MTMEM16F F518H Mutant in Digitonin within 5.0Å range:
|
Calcium binding site 6 out of 6 in 8b8j
Go back to
Calcium binding site 6 out
of 6 in the Cryo-Em Structure of CA2+-Bound MTMEM16F F518H Mutant in Digitonin

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 6 of Cryo-Em Structure of CA2+-Bound MTMEM16F F518H Mutant in Digitonin within 5.0Å range:
|
Reference:
M.Arndt,
C.Alvadia,
M.S.Straub,
V.Clerico Mosina,
C.Paulino,
R.Dutzler.
Structural Basis For the Activation of the Lipid Scramblase TMEM16F. Nat Commun V. 13 6692 2022.
ISSN: ESSN 2041-1723
PubMed: 36335104
DOI: 10.1038/S41467-022-34497-X
Page generated: Thu Jul 10 03:23:19 2025
ISSN: ESSN 2041-1723
PubMed: 36335104
DOI: 10.1038/S41467-022-34497-X
Last articles
Cl in 8BIGCl in 8BIB
Cl in 8BIF
Cl in 8BHK
Cl in 8BI6
Cl in 8BI5
Cl in 8BH0
Cl in 8BGX
Cl in 8BEM
Cl in 8BGY