Calcium »
PDB 8ban-8bsc »
8bgw »
Calcium in PDB 8bgw: Cryoem Structure of Quinol-Dependent Nitric Oxide Reductase (Qnor) From Alcaligenes Xylosoxidans at 2.2 A Resolution
Enzymatic activity of Cryoem Structure of Quinol-Dependent Nitric Oxide Reductase (Qnor) From Alcaligenes Xylosoxidans at 2.2 A Resolution
All present enzymatic activity of Cryoem Structure of Quinol-Dependent Nitric Oxide Reductase (Qnor) From Alcaligenes Xylosoxidans at 2.2 A Resolution:
1.7.2.5;
1.7.2.5;
Other elements in 8bgw:
The structure of Cryoem Structure of Quinol-Dependent Nitric Oxide Reductase (Qnor) From Alcaligenes Xylosoxidans at 2.2 A Resolution also contains other interesting chemical elements:
| Iron | (Fe) | 6 atoms |
Calcium Binding Sites:
The binding sites of Calcium atom in the Cryoem Structure of Quinol-Dependent Nitric Oxide Reductase (Qnor) From Alcaligenes Xylosoxidans at 2.2 A Resolution
(pdb code 8bgw). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Cryoem Structure of Quinol-Dependent Nitric Oxide Reductase (Qnor) From Alcaligenes Xylosoxidans at 2.2 A Resolution, PDB code: 8bgw:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Cryoem Structure of Quinol-Dependent Nitric Oxide Reductase (Qnor) From Alcaligenes Xylosoxidans at 2.2 A Resolution, PDB code: 8bgw:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 8bgw
Go back to
Calcium binding site 1 out
of 2 in the Cryoem Structure of Quinol-Dependent Nitric Oxide Reductase (Qnor) From Alcaligenes Xylosoxidans at 2.2 A Resolution
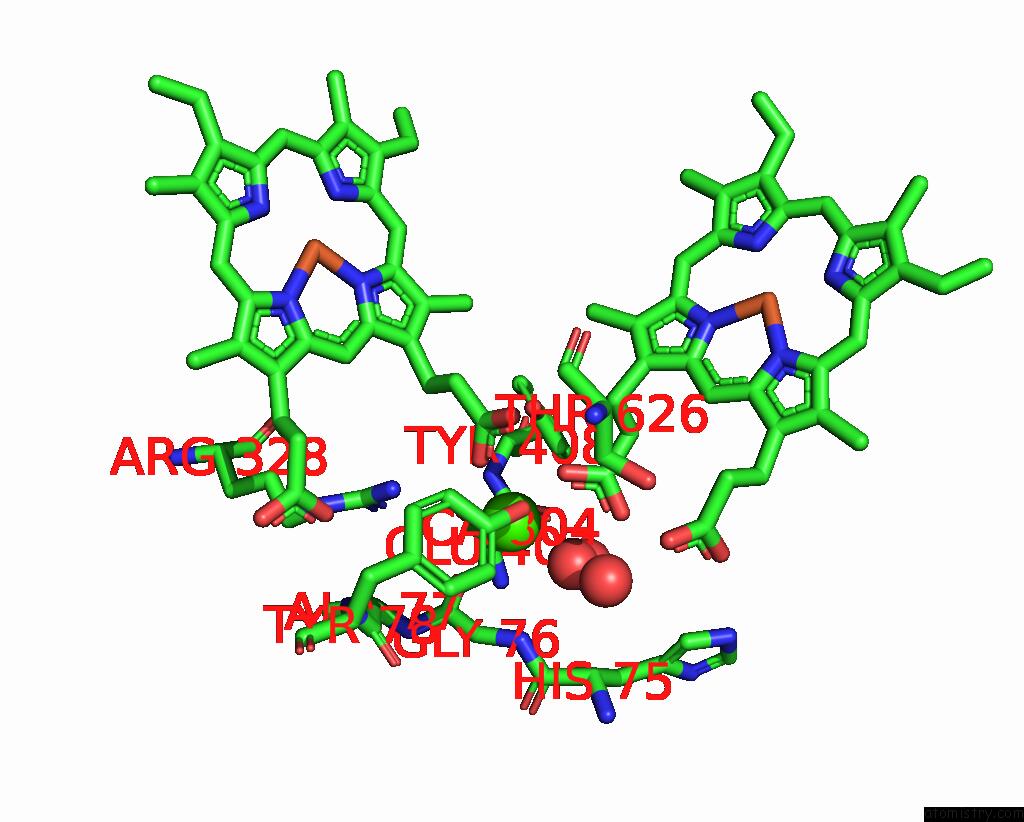
Mono view
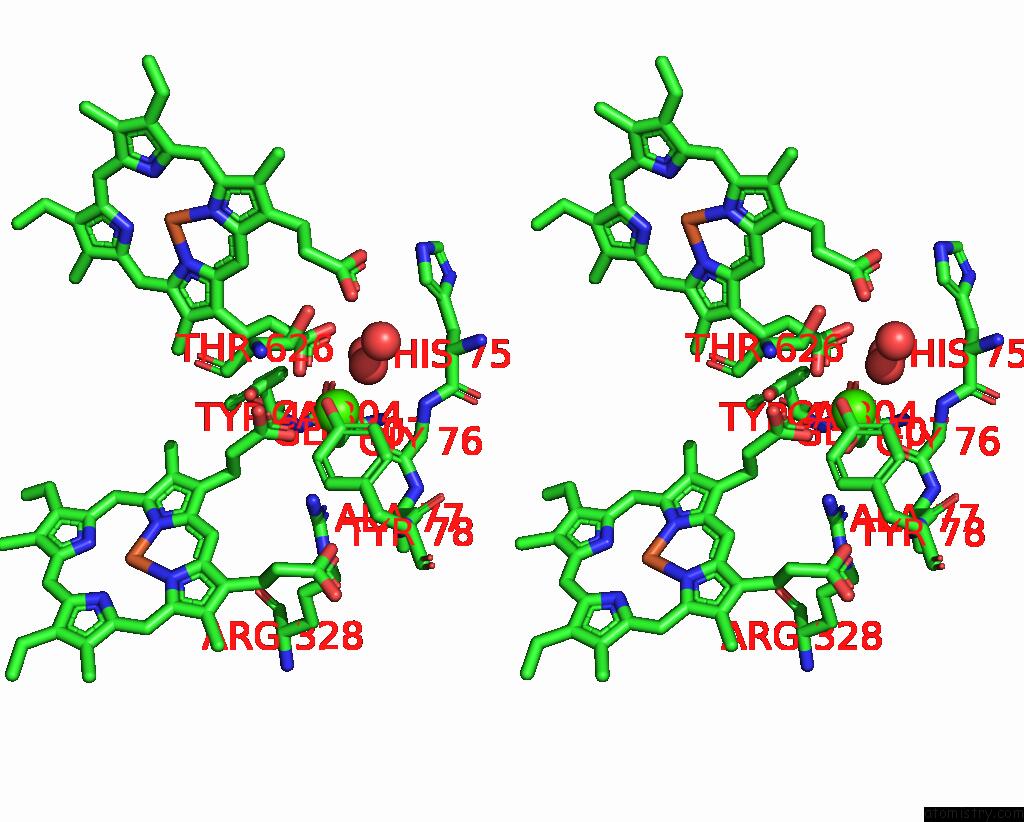
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Cryoem Structure of Quinol-Dependent Nitric Oxide Reductase (Qnor) From Alcaligenes Xylosoxidans at 2.2 A Resolution within 5.0Å range:
|
Calcium binding site 2 out of 2 in 8bgw
Go back to
Calcium binding site 2 out
of 2 in the Cryoem Structure of Quinol-Dependent Nitric Oxide Reductase (Qnor) From Alcaligenes Xylosoxidans at 2.2 A Resolution
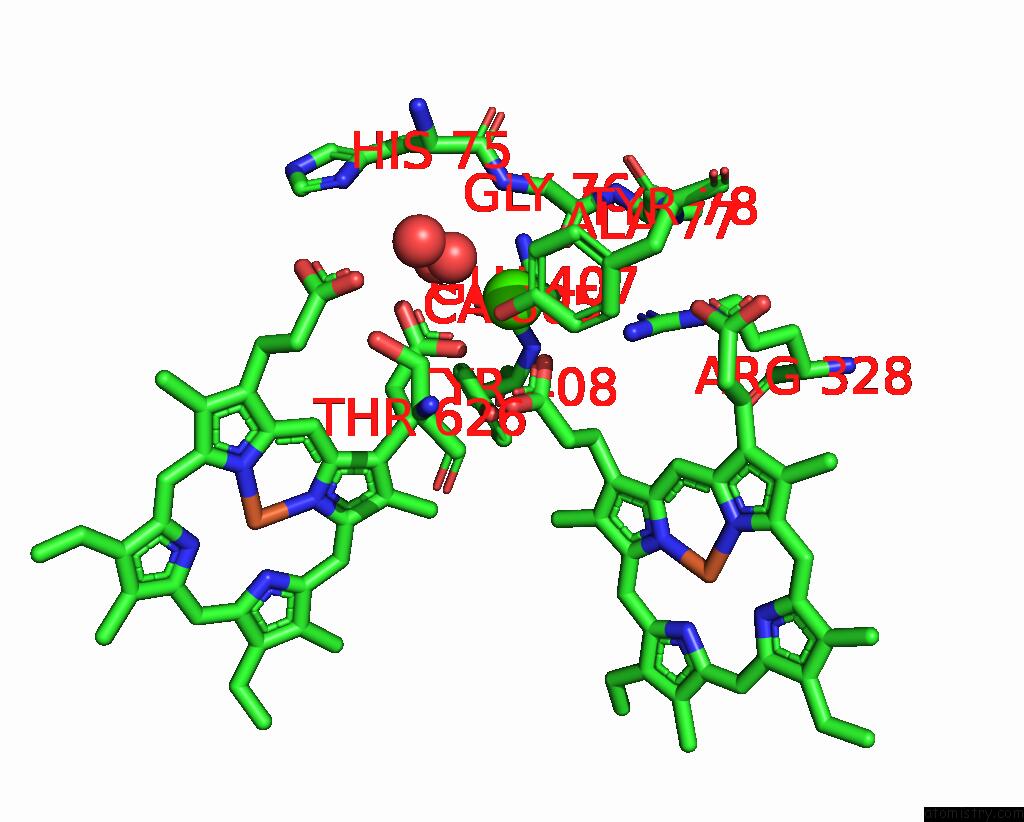
Mono view
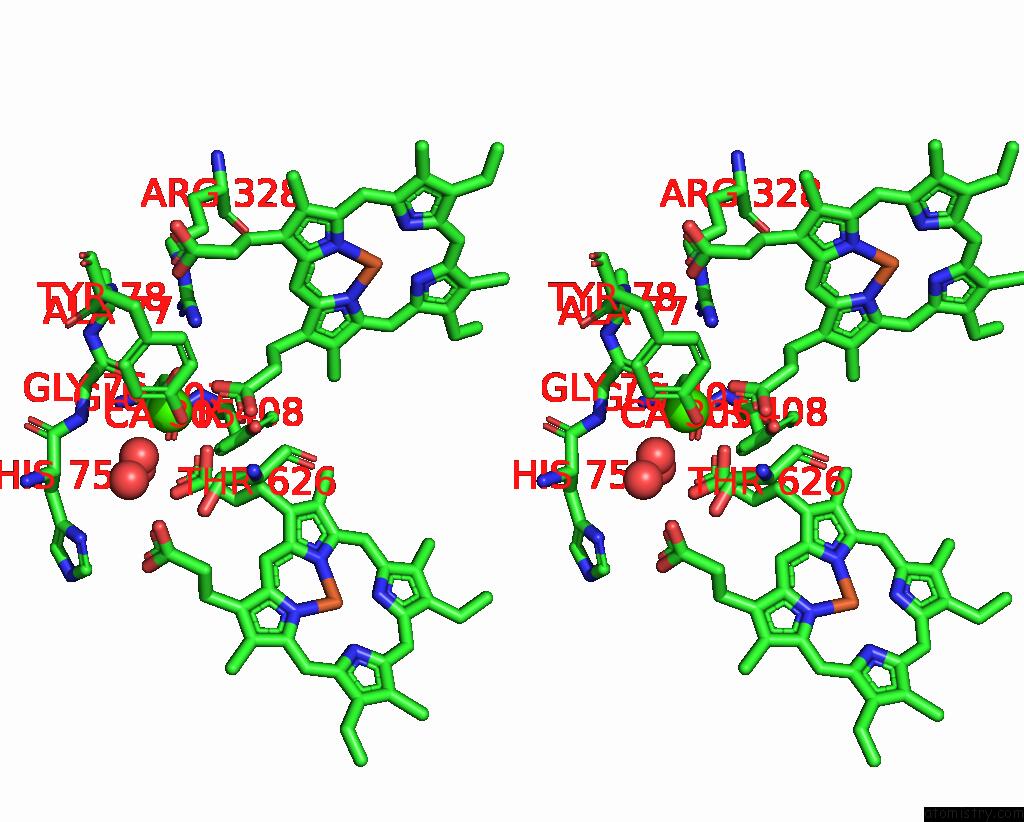
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Cryoem Structure of Quinol-Dependent Nitric Oxide Reductase (Qnor) From Alcaligenes Xylosoxidans at 2.2 A Resolution within 5.0Å range:
|
Reference:
A.J.Flynn,
S.V.Antonyuk,
R.R.Eady,
S.P.Muench,
S.S.Hasnain.
A 2.2 Angstrom Cryoem Structure of A Quinol-Dependent No Reductase Shows Close Similarity to Respiratory Oxidases. Nat Commun V. 14 3416 2023.
ISSN: ESSN 2041-1723
PubMed: 37296134
DOI: 10.1038/S41467-023-39140-X
Page generated: Thu Jul 10 03:29:30 2025
ISSN: ESSN 2041-1723
PubMed: 37296134
DOI: 10.1038/S41467-023-39140-X
Last articles
Cl in 5J2BCl in 5J2A
Cl in 5IXL
Cl in 5J0Z
Cl in 5J1T
Cl in 5J1S
Cl in 5IXY
Cl in 5IXE
Cl in 5IXS
Cl in 5IVT