Calcium »
PDB 8ban-8bsc »
8brq »
Calcium in PDB 8brq: Crystal Structure of A Surface Entropy Reduction Variant of Penicillin G Acylase From Bacillaceae I. S. Sp. Fjat-27231
Protein crystallography data
The structure of Crystal Structure of A Surface Entropy Reduction Variant of Penicillin G Acylase From Bacillaceae I. S. Sp. Fjat-27231, PDB code: 8brq
was solved by
J.Wichmann,
J.Mayer,
H.Mattes,
P.Lukat,
W.Blankenfeldt,
R.Biedendieck,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 52.53 / 1.63 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 61.252, 102.105, 140.079, 90, 90, 90 |
| R / Rfree (%) | 14.3 / 16.8 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of A Surface Entropy Reduction Variant of Penicillin G Acylase From Bacillaceae I. S. Sp. Fjat-27231
(pdb code 8brq). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Crystal Structure of A Surface Entropy Reduction Variant of Penicillin G Acylase From Bacillaceae I. S. Sp. Fjat-27231, PDB code: 8brq:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Crystal Structure of A Surface Entropy Reduction Variant of Penicillin G Acylase From Bacillaceae I. S. Sp. Fjat-27231, PDB code: 8brq:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 8brq
Go back to
Calcium binding site 1 out
of 2 in the Crystal Structure of A Surface Entropy Reduction Variant of Penicillin G Acylase From Bacillaceae I. S. Sp. Fjat-27231

Mono view
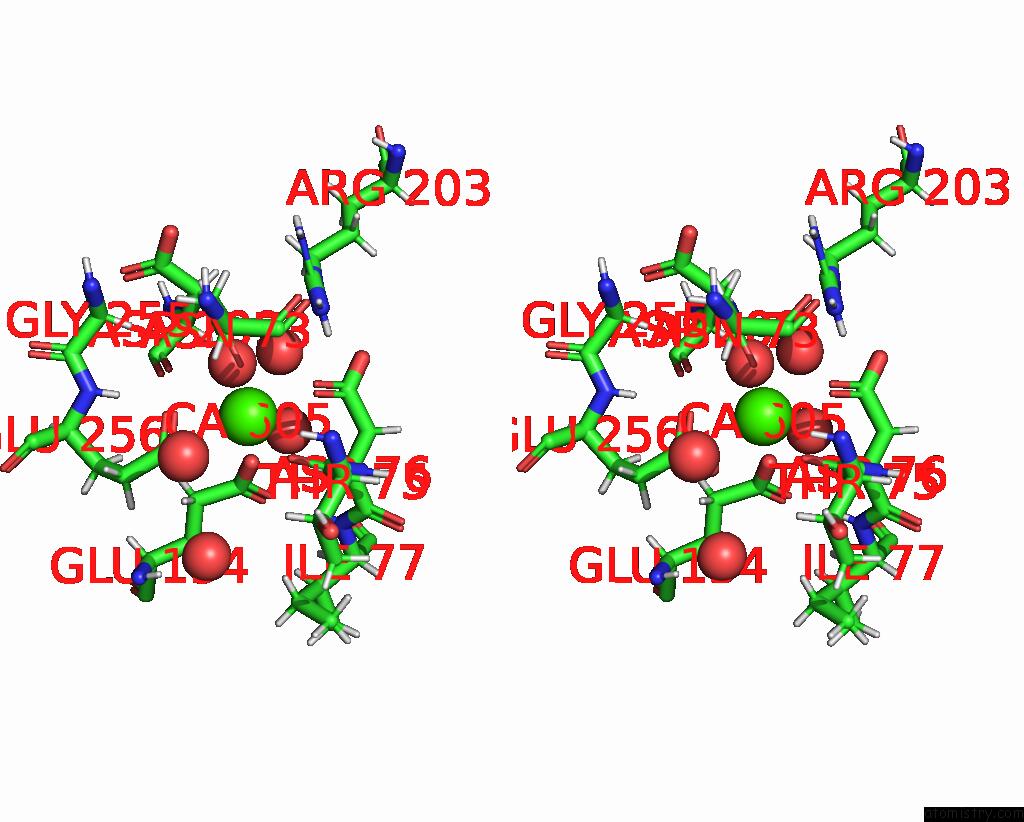
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of A Surface Entropy Reduction Variant of Penicillin G Acylase From Bacillaceae I. S. Sp. Fjat-27231 within 5.0Å range:
|
Calcium binding site 2 out of 2 in 8brq
Go back to
Calcium binding site 2 out
of 2 in the Crystal Structure of A Surface Entropy Reduction Variant of Penicillin G Acylase From Bacillaceae I. S. Sp. Fjat-27231
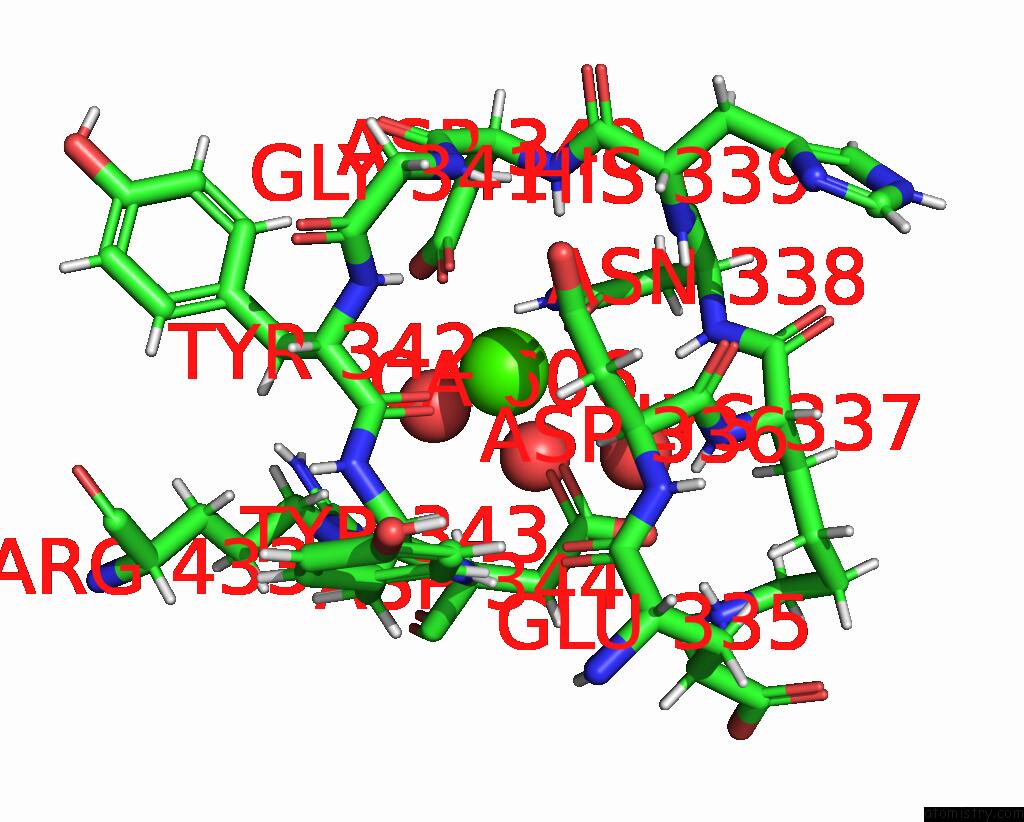
Mono view
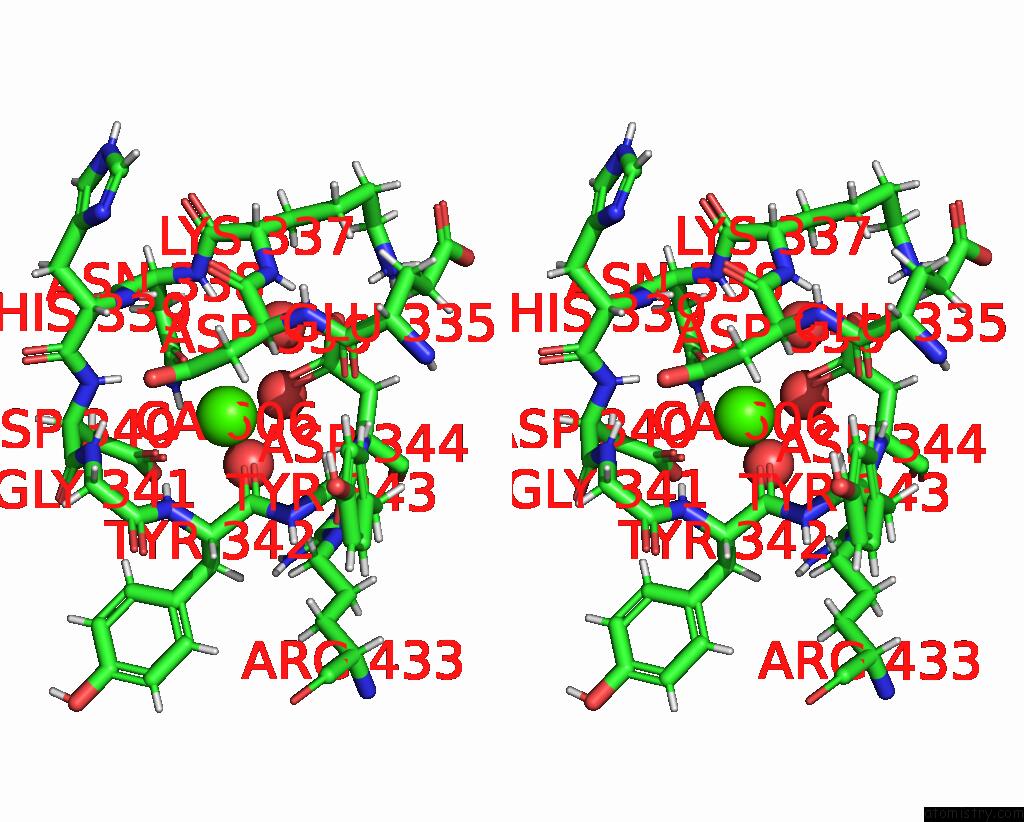
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Crystal Structure of A Surface Entropy Reduction Variant of Penicillin G Acylase From Bacillaceae I. S. Sp. Fjat-27231 within 5.0Å range:
|
Reference:
J.Wichmann,
J.Mayer,
M.Hintmann,
P.Lukat,
W.Blankenfeldt,
R.Biedendieck.
Multistep Engineering of A Penicillin G Acylase For Systematic Improvement of Crystallization Efficiency Cryst.Growth Des. 2023.
ISSN: ESSN 1528-7505
DOI: 10.1021/ACS.CGD.2C01408
Page generated: Thu Jul 10 03:33:37 2025
ISSN: ESSN 1528-7505
DOI: 10.1021/ACS.CGD.2C01408
Last articles
Cl in 5R9CCl in 5R9E
Cl in 5R9B
Cl in 5R9D
Cl in 5R9A
Cl in 5R99
Cl in 5R98
Cl in 5R96
Cl in 5R97
Cl in 5R95