Calcium »
PDB 8fg4-8fvp »
8fji »
Calcium in PDB 8fji: Crystal Structure of Rala in A Covalent Complex with Sof-367
Protein crystallography data
The structure of Crystal Structure of Rala in A Covalent Complex with Sof-367, PDB code: 8fji
was solved by
A.D.Landgraf,
I.-J.Yeh,
K.Bum-Erdene,
G.Gonzalez-Gutierrez,
S.Meroueh,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 55.07 / 1.48 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 64.767, 104.63, 55.823, 90, 90, 90 |
| R / Rfree (%) | 18.3 / 20.9 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of Rala in A Covalent Complex with Sof-367
(pdb code 8fji). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 3 binding sites of Calcium where determined in the Crystal Structure of Rala in A Covalent Complex with Sof-367, PDB code: 8fji:
Jump to Calcium binding site number: 1; 2; 3;
In total 3 binding sites of Calcium where determined in the Crystal Structure of Rala in A Covalent Complex with Sof-367, PDB code: 8fji:
Jump to Calcium binding site number: 1; 2; 3;
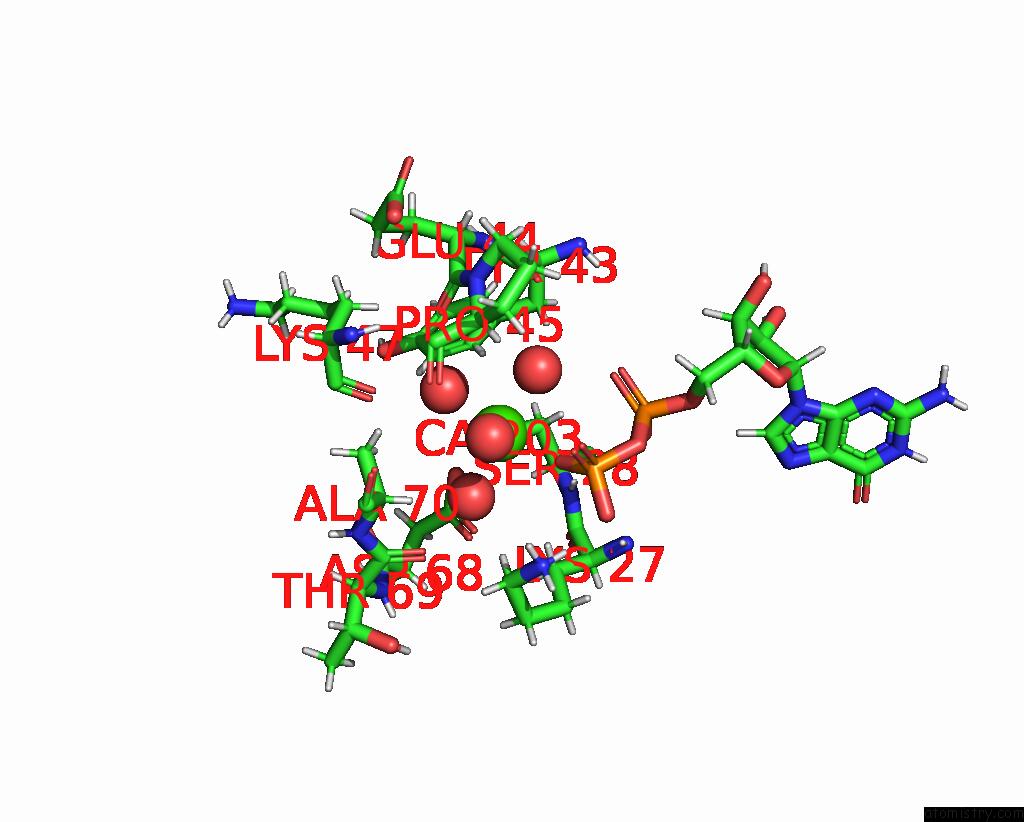
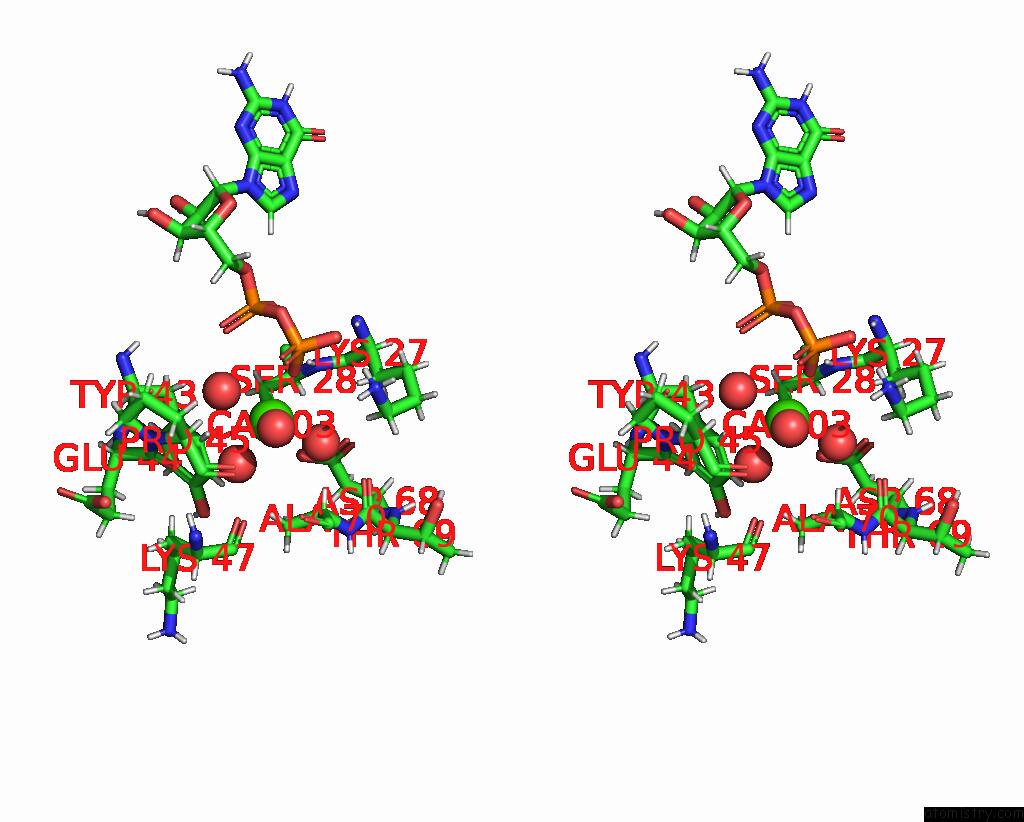
Calcium binding site 1 out of 3 in 8fji
Go back to
Calcium binding site 1 out
of 3 in the Crystal Structure of Rala in A Covalent Complex with Sof-367
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of Rala in A Covalent Complex with Sof-367 within 5.0Å range:
|
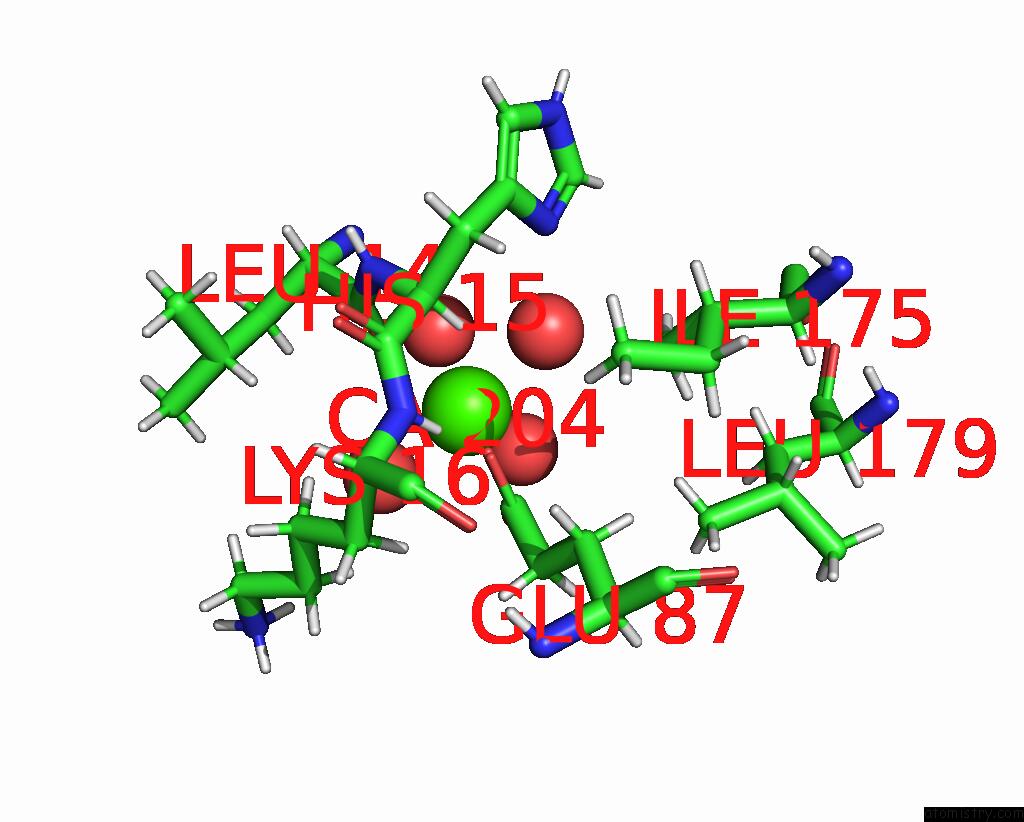
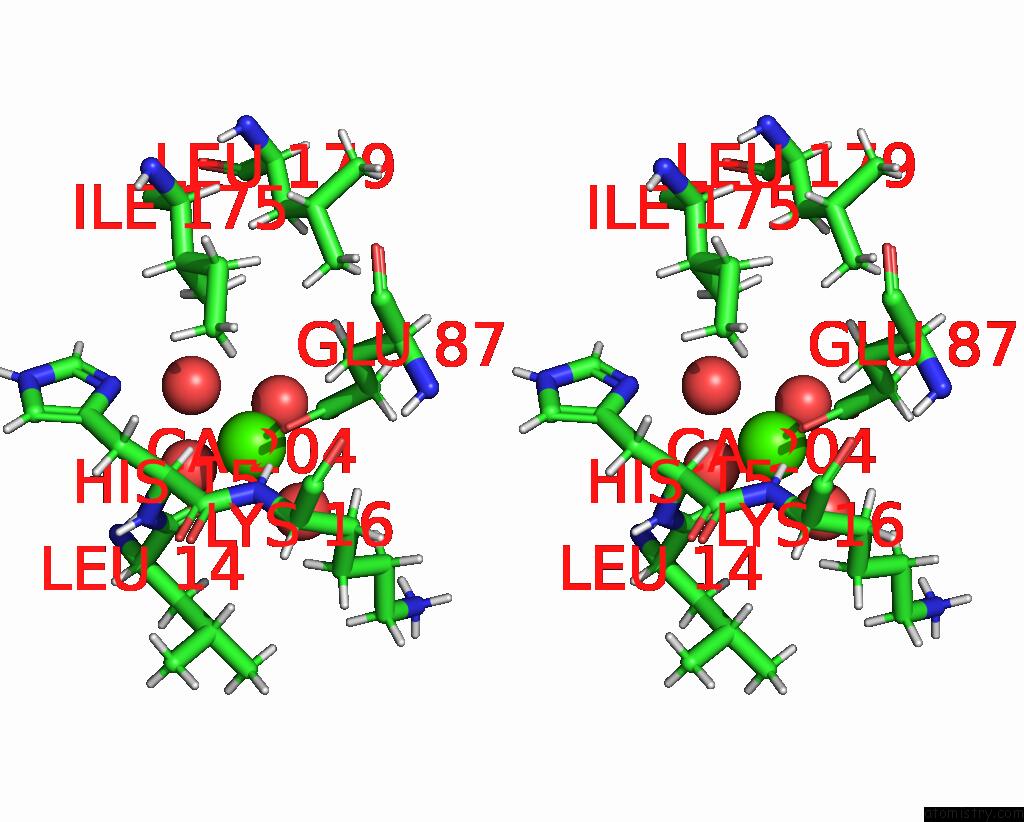
Calcium binding site 2 out of 3 in 8fji
Go back to
Calcium binding site 2 out
of 3 in the Crystal Structure of Rala in A Covalent Complex with Sof-367
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Crystal Structure of Rala in A Covalent Complex with Sof-367 within 5.0Å range:
|
Calcium binding site 3 out of 3 in 8fji
Go back to
Calcium binding site 3 out
of 3 in the Crystal Structure of Rala in A Covalent Complex with Sof-367
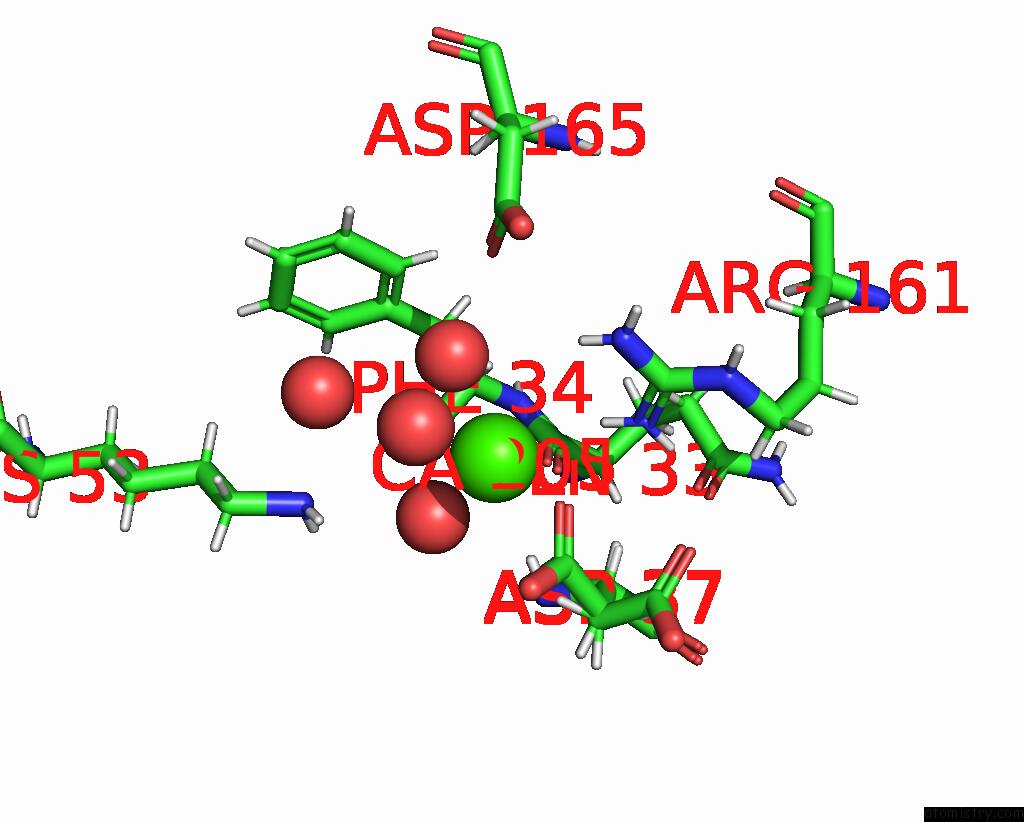
Mono view
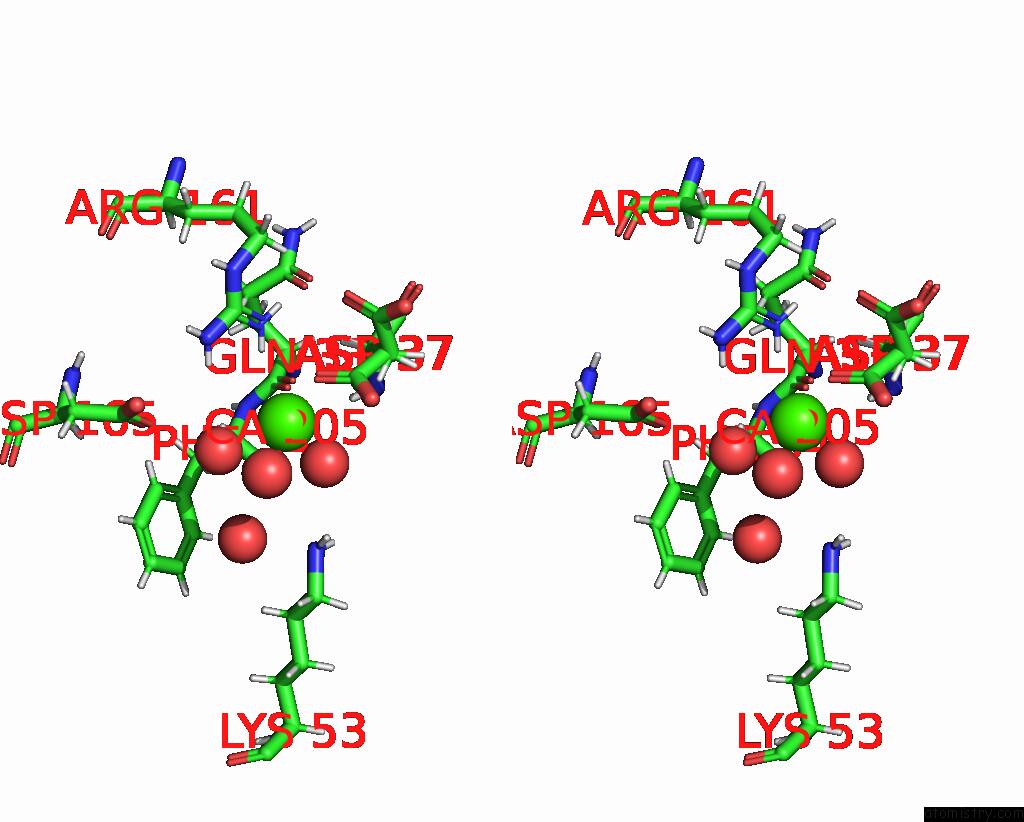
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Crystal Structure of Rala in A Covalent Complex with Sof-367 within 5.0Å range:
|
Reference:
A.D.Landgraf,
I.J.Yeh,
M.K.Ghozayel,
K.Bum-Erdene,
G.Gonzalez-Gutierrez,
S.O.Meroueh.
Exploring Covalent Bond Formation at Tyr-82 For Inhibition of Ral Gtpase Activation. Chemmedchem V. 18 00272 2023.
ISSN: ESSN 1860-7187
PubMed: 37269475
DOI: 10.1002/CMDC.202300272
Page generated: Thu Jul 10 04:40:20 2025
ISSN: ESSN 1860-7187
PubMed: 37269475
DOI: 10.1002/CMDC.202300272
Last articles
Cd in 3ZI7Cd in 3ZG5
Cd in 3ZFZ
Cd in 3ZG0
Cd in 3WZO
Cd in 3ZET
Cd in 3WNK
Cd in 3X1Y
Cd in 3X1X
Cd in 3X1W