Calcium »
PDB 8olc-8q29 »
8pe9 »
Calcium in PDB 8pe9: Complex Between DDR1 Ds-Like Domain and Prth-101 Fab
Enzymatic activity of Complex Between DDR1 Ds-Like Domain and Prth-101 Fab
All present enzymatic activity of Complex Between DDR1 Ds-Like Domain and Prth-101 Fab:
2.7.10.1;
2.7.10.1;
Protein crystallography data
The structure of Complex Between DDR1 Ds-Like Domain and Prth-101 Fab, PDB code: 8pe9
was solved by
J.Liu,
H.Chiang,
W.Xiong,
V.Laurent,
S.C.Griffiths,
J.Duelfer,
H.Deng,
X.Sun,
Y.W.Yin,
W.Li,
L.P.Audoly,
Z.An,
T.Schuerpf,
R.Li,
N.Zhang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 72.22 / 3.15 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 117.096, 144.45, 52.299, 90, 90, 90 |
| R / Rfree (%) | 27.8 / 30.9 |
Other elements in 8pe9:
The structure of Complex Between DDR1 Ds-Like Domain and Prth-101 Fab also contains other interesting chemical elements:
| Zinc | (Zn) | 3 atoms |
| Sodium | (Na) | 1 atom |
Calcium Binding Sites:
The binding sites of Calcium atom in the Complex Between DDR1 Ds-Like Domain and Prth-101 Fab
(pdb code 8pe9). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total only one binding site of Calcium was determined in the Complex Between DDR1 Ds-Like Domain and Prth-101 Fab, PDB code: 8pe9:
In total only one binding site of Calcium was determined in the Complex Between DDR1 Ds-Like Domain and Prth-101 Fab, PDB code: 8pe9:
Calcium binding site 1 out of 1 in 8pe9
Go back to
Calcium binding site 1 out
of 1 in the Complex Between DDR1 Ds-Like Domain and Prth-101 Fab

Mono view
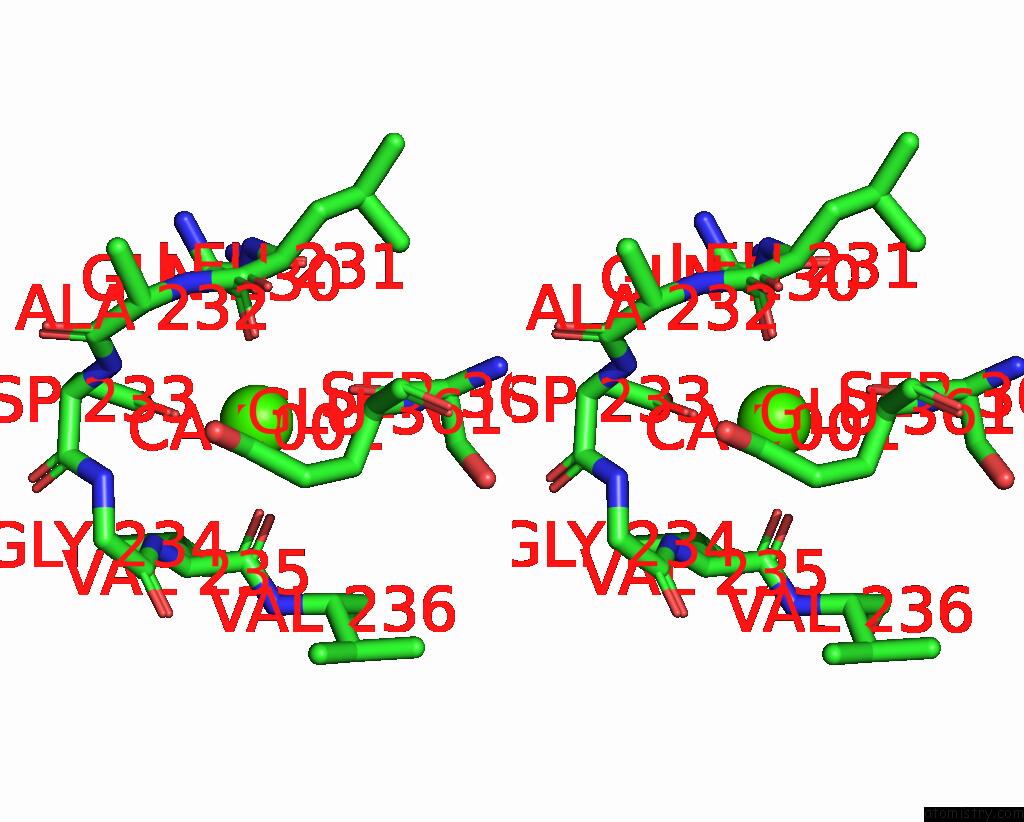
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Complex Between DDR1 Ds-Like Domain and Prth-101 Fab within 5.0Å range:
|
Reference:
J.Liu,
H.C.Chiang,
W.Xiong,
V.Laurent,
S.C.Griffiths,
J.Dulfer,
H.Deng,
X.Sun,
Y.W.Yin,
W.Li,
L.P.Audoly,
Z.An,
T.Schurpf,
R.Li,
N.Zhang.
A Highly Selective Humanized DDR1 Mab Reverses Immune Exclusion By Disrupting Collagen Fiber Alignment in Breast Cancer. J Immunother Cancer V. 11 2023.
ISSN: ESSN 2051-1426
PubMed: 37328286
DOI: 10.1136/JITC-2023-006720
Page generated: Thu Jul 10 06:27:18 2025
ISSN: ESSN 2051-1426
PubMed: 37328286
DOI: 10.1136/JITC-2023-006720
Last articles
Cl in 8CCBCl in 8CCD
Cl in 8CCC
Cl in 8CC6
Cl in 8CBH
Cl in 8CBV
Cl in 8CBU
Cl in 8CB6
Cl in 8CBT
Cl in 8CBS