Calcium »
PDB 8szi-8tqb »
8tpo »
Calcium in PDB 8tpo: NHTMEM16 R432A Mutant in Lipid Nanodiscs with MSP1E3 Scaffold Protein in the Presence of CA2+
Calcium Binding Sites:
The binding sites of Calcium atom in the NHTMEM16 R432A Mutant in Lipid Nanodiscs with MSP1E3 Scaffold Protein in the Presence of CA2+
(pdb code 8tpo). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 4 binding sites of Calcium where determined in the NHTMEM16 R432A Mutant in Lipid Nanodiscs with MSP1E3 Scaffold Protein in the Presence of CA2+, PDB code: 8tpo:
Jump to Calcium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Calcium where determined in the NHTMEM16 R432A Mutant in Lipid Nanodiscs with MSP1E3 Scaffold Protein in the Presence of CA2+, PDB code: 8tpo:
Jump to Calcium binding site number: 1; 2; 3; 4;
Calcium binding site 1 out of 4 in 8tpo
Go back to
Calcium binding site 1 out
of 4 in the NHTMEM16 R432A Mutant in Lipid Nanodiscs with MSP1E3 Scaffold Protein in the Presence of CA2+
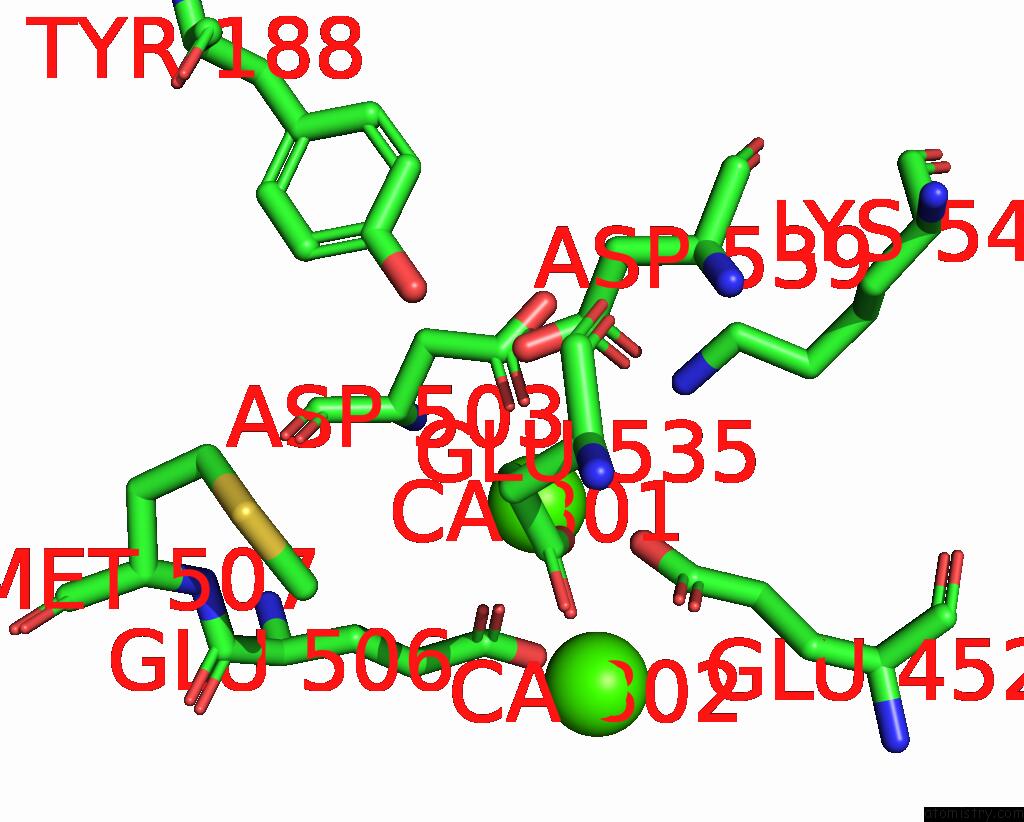
Mono view
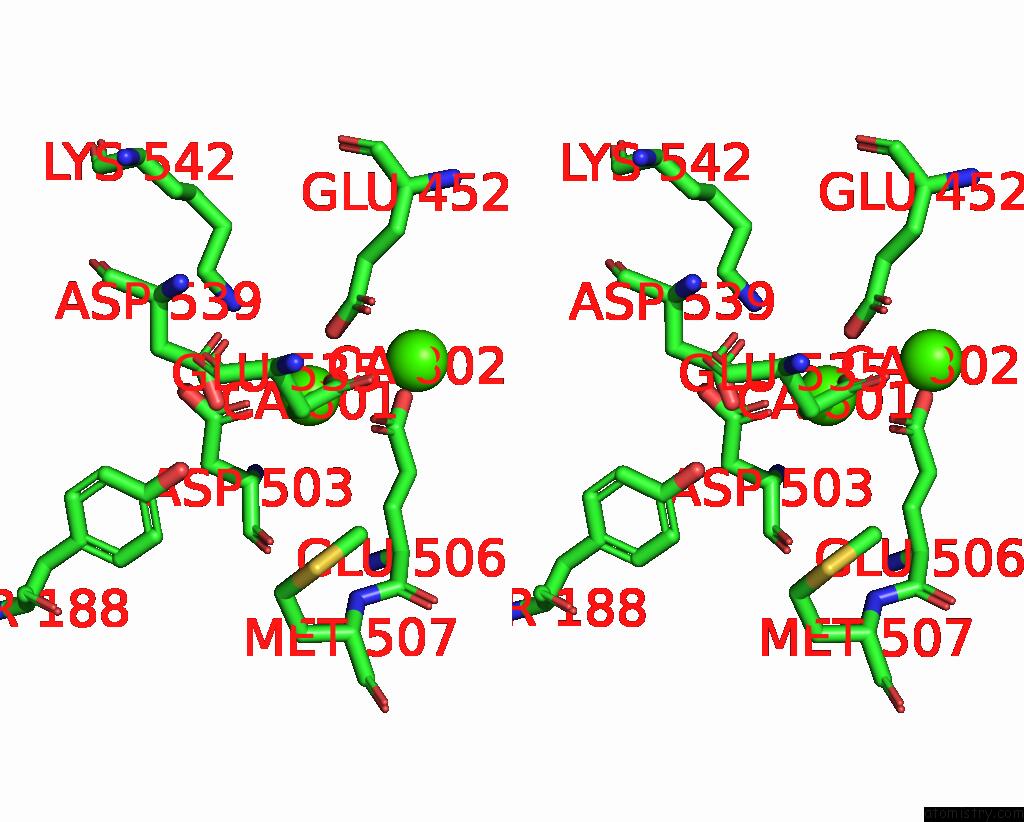
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of NHTMEM16 R432A Mutant in Lipid Nanodiscs with MSP1E3 Scaffold Protein in the Presence of CA2+ within 5.0Å range:
|
Calcium binding site 2 out of 4 in 8tpo
Go back to
Calcium binding site 2 out
of 4 in the NHTMEM16 R432A Mutant in Lipid Nanodiscs with MSP1E3 Scaffold Protein in the Presence of CA2+
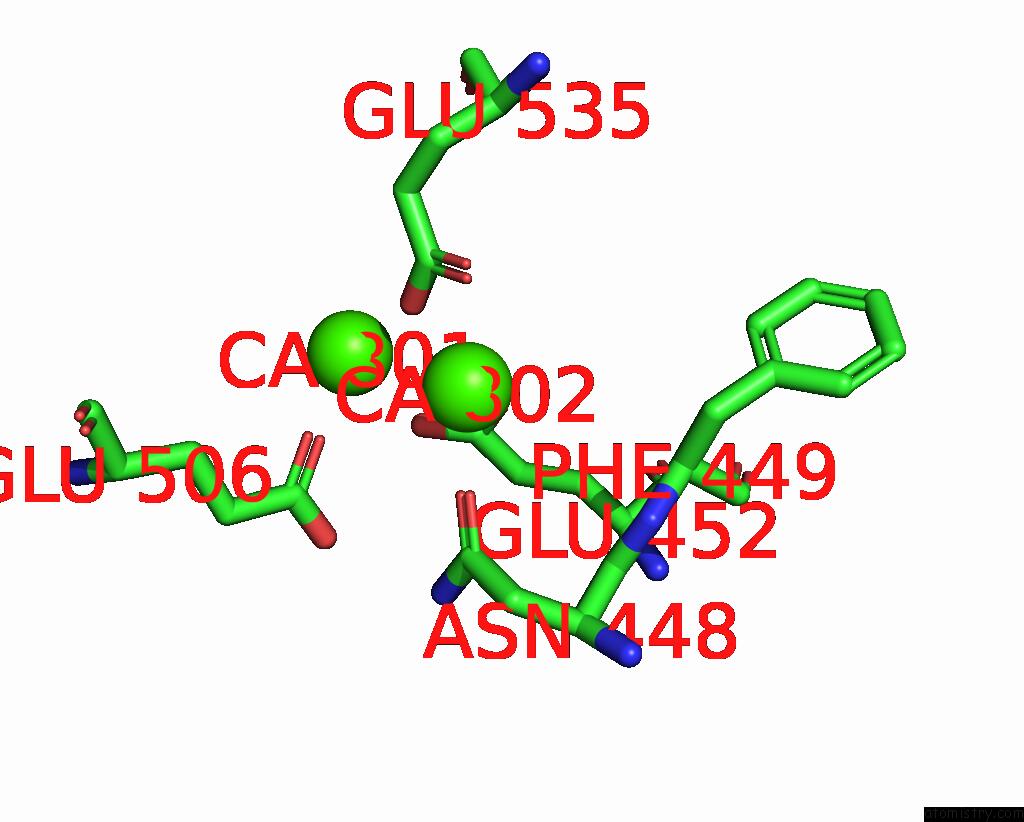
Mono view
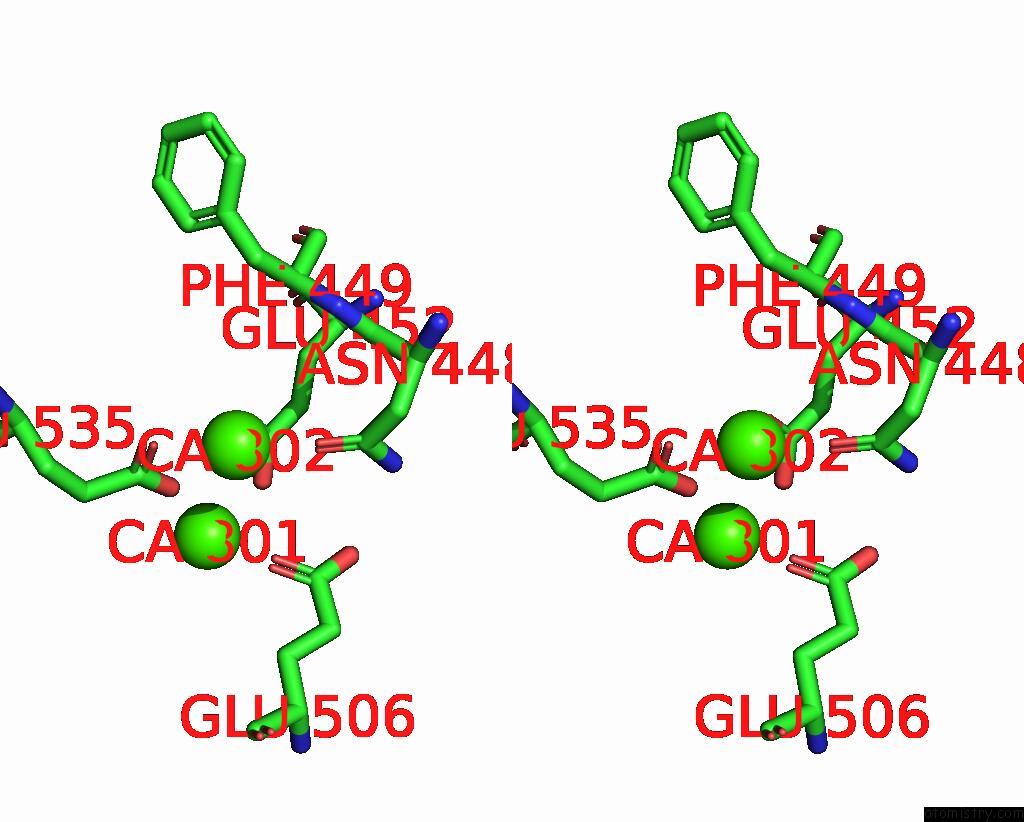
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of NHTMEM16 R432A Mutant in Lipid Nanodiscs with MSP1E3 Scaffold Protein in the Presence of CA2+ within 5.0Å range:
|
Calcium binding site 3 out of 4 in 8tpo
Go back to
Calcium binding site 3 out
of 4 in the NHTMEM16 R432A Mutant in Lipid Nanodiscs with MSP1E3 Scaffold Protein in the Presence of CA2+
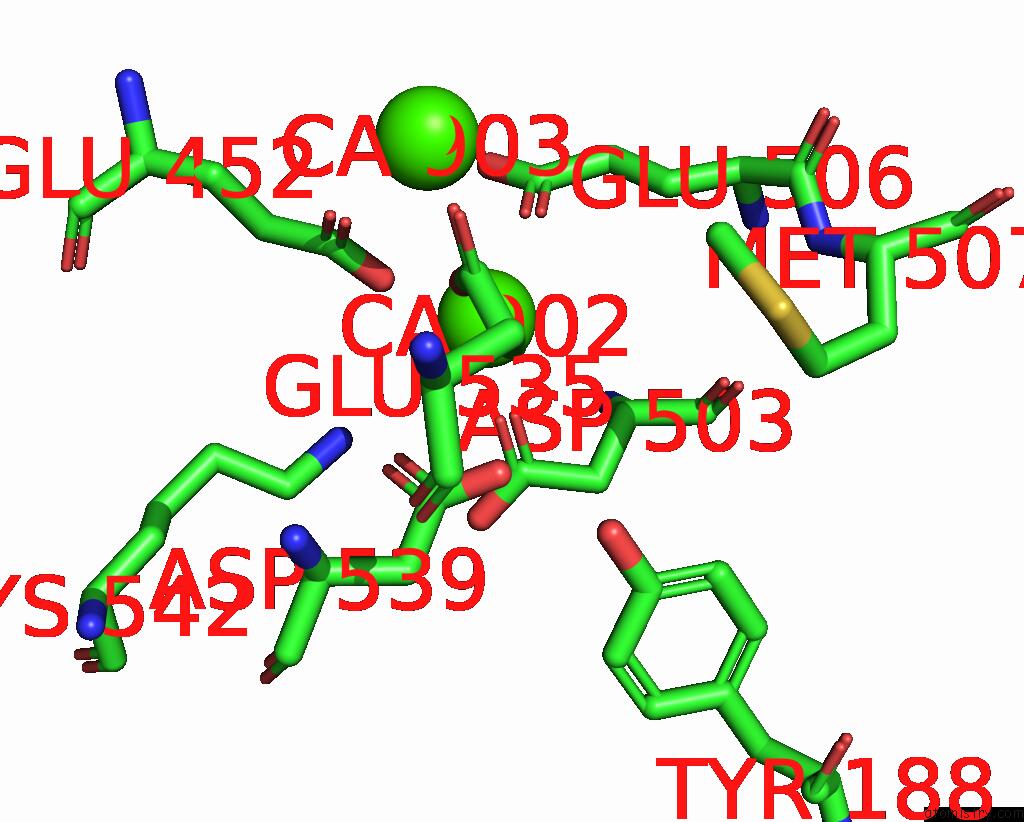
Mono view
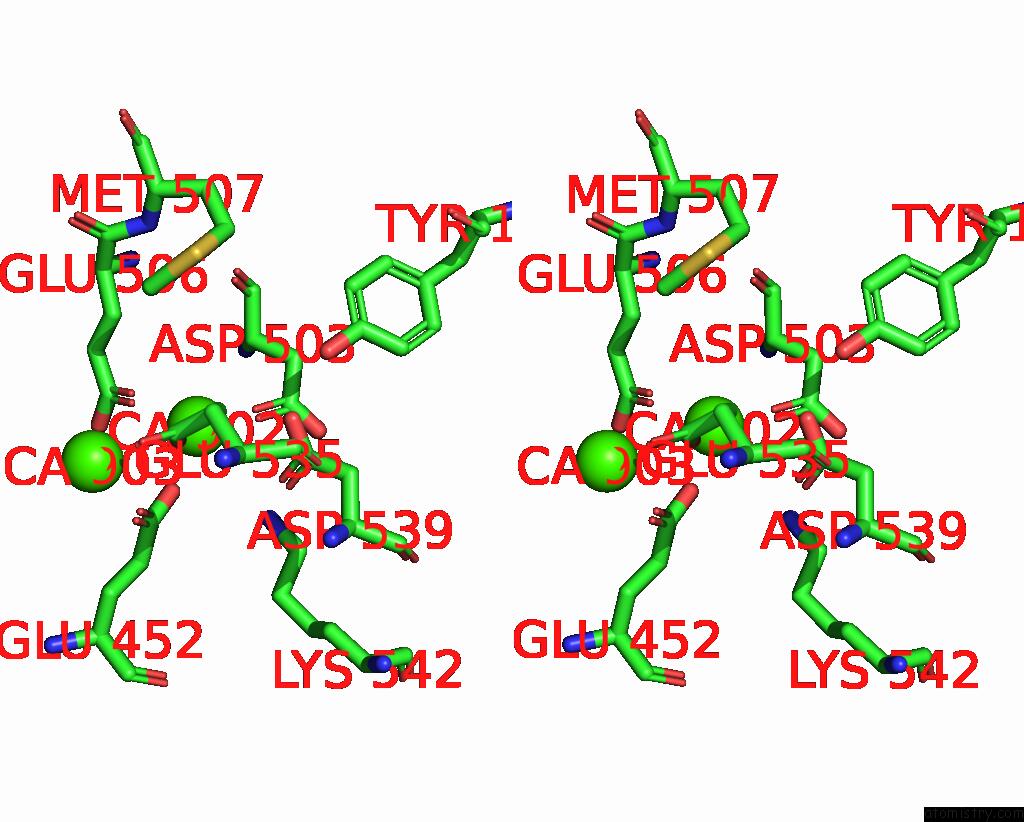
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of NHTMEM16 R432A Mutant in Lipid Nanodiscs with MSP1E3 Scaffold Protein in the Presence of CA2+ within 5.0Å range:
|
Calcium binding site 4 out of 4 in 8tpo
Go back to
Calcium binding site 4 out
of 4 in the NHTMEM16 R432A Mutant in Lipid Nanodiscs with MSP1E3 Scaffold Protein in the Presence of CA2+
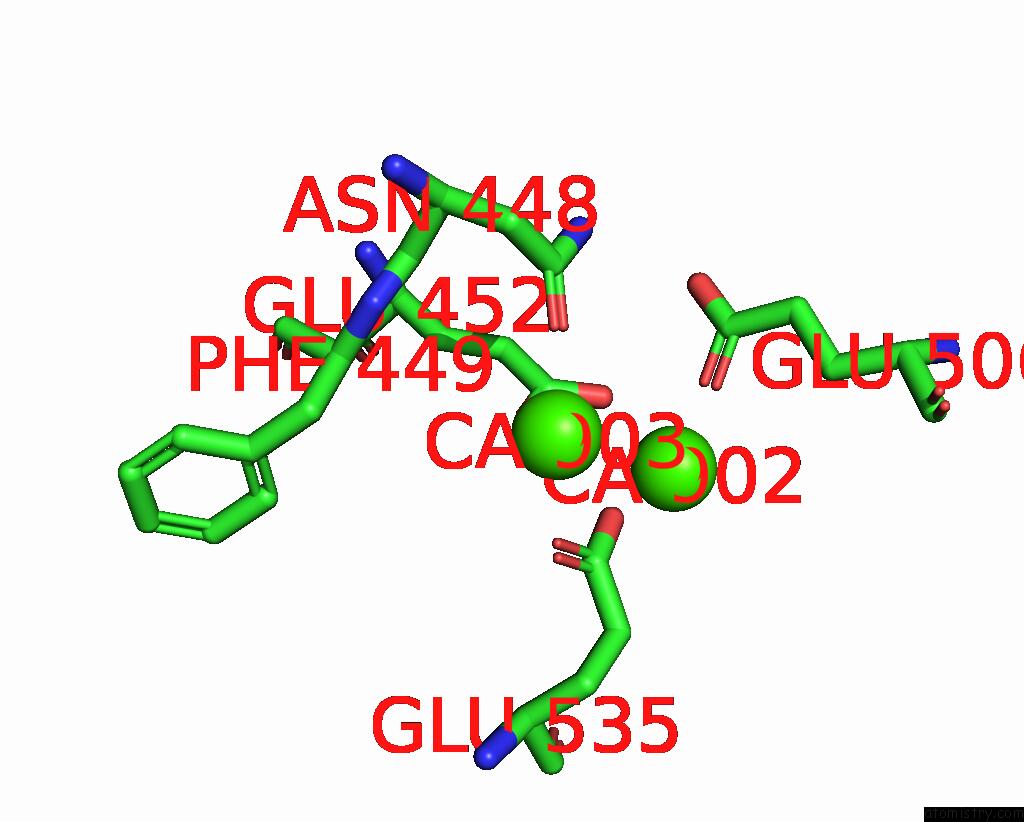
Mono view
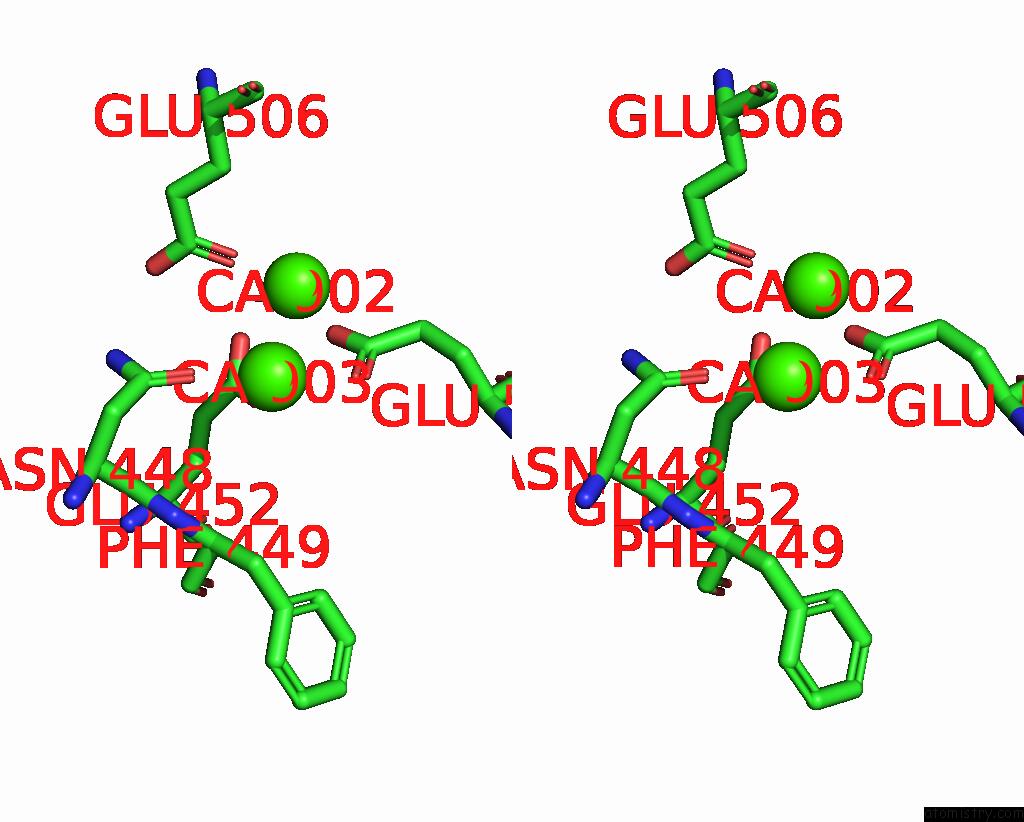
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of NHTMEM16 R432A Mutant in Lipid Nanodiscs with MSP1E3 Scaffold Protein in the Presence of CA2+ within 5.0Å range:
|
Reference:
Z.Feng,
O.E.Alvarenga,
A.Accardi.
Structural Basis of Closed Groove Scrambling By A TMEM16 Protein. Nat.Struct.Mol.Biol. 2024.
ISSN: ESSN 1545-9985
PubMed: 38684930
DOI: 10.1038/S41594-024-01284-9
Page generated: Thu Jul 10 07:33:54 2025
ISSN: ESSN 1545-9985
PubMed: 38684930
DOI: 10.1038/S41594-024-01284-9
Last articles
F in 4G1WF in 4G16
F in 4FXY
F in 4FXQ
F in 4FZF
F in 4FP1
F in 4FVX
F in 4FV1
F in 4FV9
F in 4FV3