Calcium »
PDB 8tr2-8uwm »
8urf »
Calcium in PDB 8urf: Crystal Structure of Human ASGR2 Crd (Carbohydrate Recognition Domain) Bound to 8G8 Fab
Protein crystallography data
The structure of Crystal Structure of Human ASGR2 Crd (Carbohydrate Recognition Domain) Bound to 8G8 Fab, PDB code: 8urf
was solved by
P.Sampathumar,
Y.Li,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 33.54 / 1.90 |
| Space group | H 3 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 102.41, 102.41, 358.985, 90, 90, 120 |
| R / Rfree (%) | 16.6 / 20.4 |
Other elements in 8urf:
The structure of Crystal Structure of Human ASGR2 Crd (Carbohydrate Recognition Domain) Bound to 8G8 Fab also contains other interesting chemical elements:
| Chlorine | (Cl) | 1 atom |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of Human ASGR2 Crd (Carbohydrate Recognition Domain) Bound to 8G8 Fab
(pdb code 8urf). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 3 binding sites of Calcium where determined in the Crystal Structure of Human ASGR2 Crd (Carbohydrate Recognition Domain) Bound to 8G8 Fab, PDB code: 8urf:
Jump to Calcium binding site number: 1; 2; 3;
In total 3 binding sites of Calcium where determined in the Crystal Structure of Human ASGR2 Crd (Carbohydrate Recognition Domain) Bound to 8G8 Fab, PDB code: 8urf:
Jump to Calcium binding site number: 1; 2; 3;
Calcium binding site 1 out of 3 in 8urf
Go back to
Calcium binding site 1 out
of 3 in the Crystal Structure of Human ASGR2 Crd (Carbohydrate Recognition Domain) Bound to 8G8 Fab
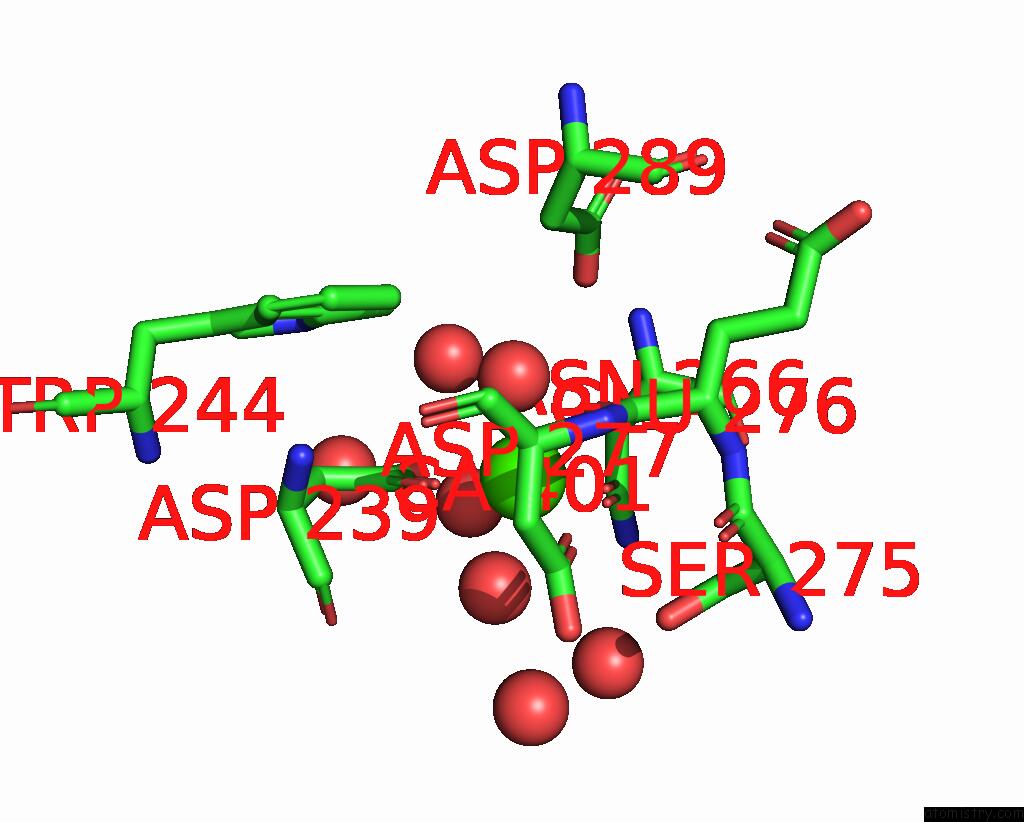
Mono view
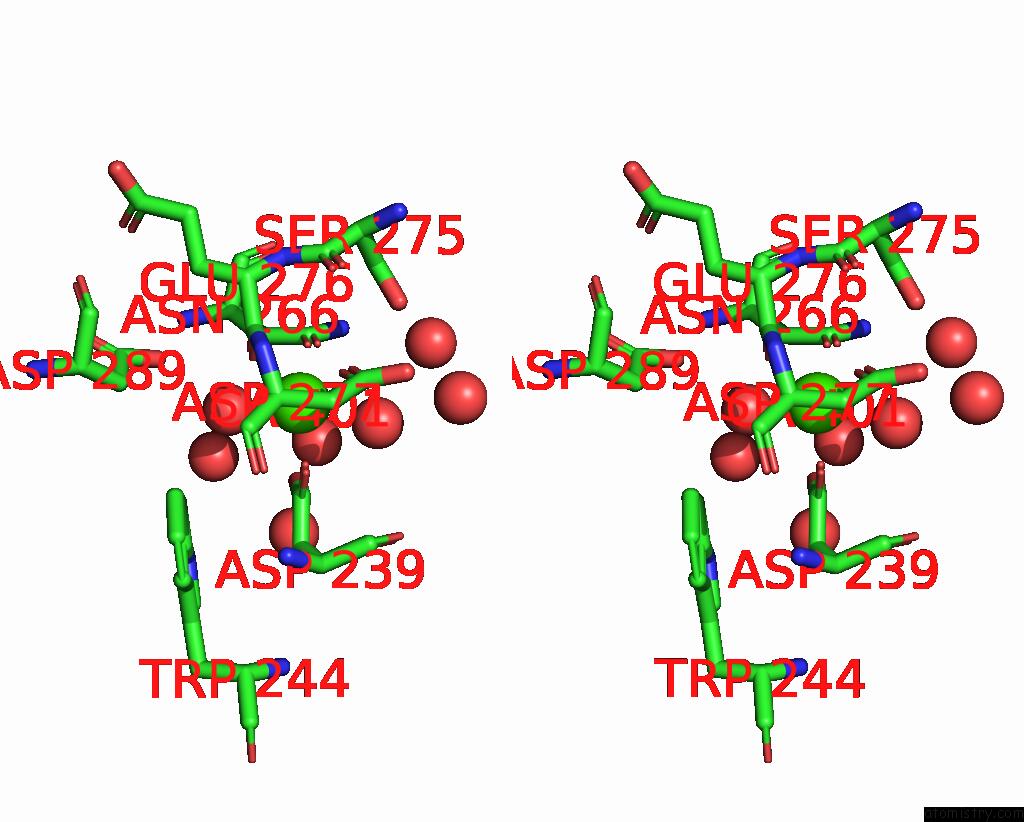
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of Human ASGR2 Crd (Carbohydrate Recognition Domain) Bound to 8G8 Fab within 5.0Å range:
|
Calcium binding site 2 out of 3 in 8urf
Go back to
Calcium binding site 2 out
of 3 in the Crystal Structure of Human ASGR2 Crd (Carbohydrate Recognition Domain) Bound to 8G8 Fab
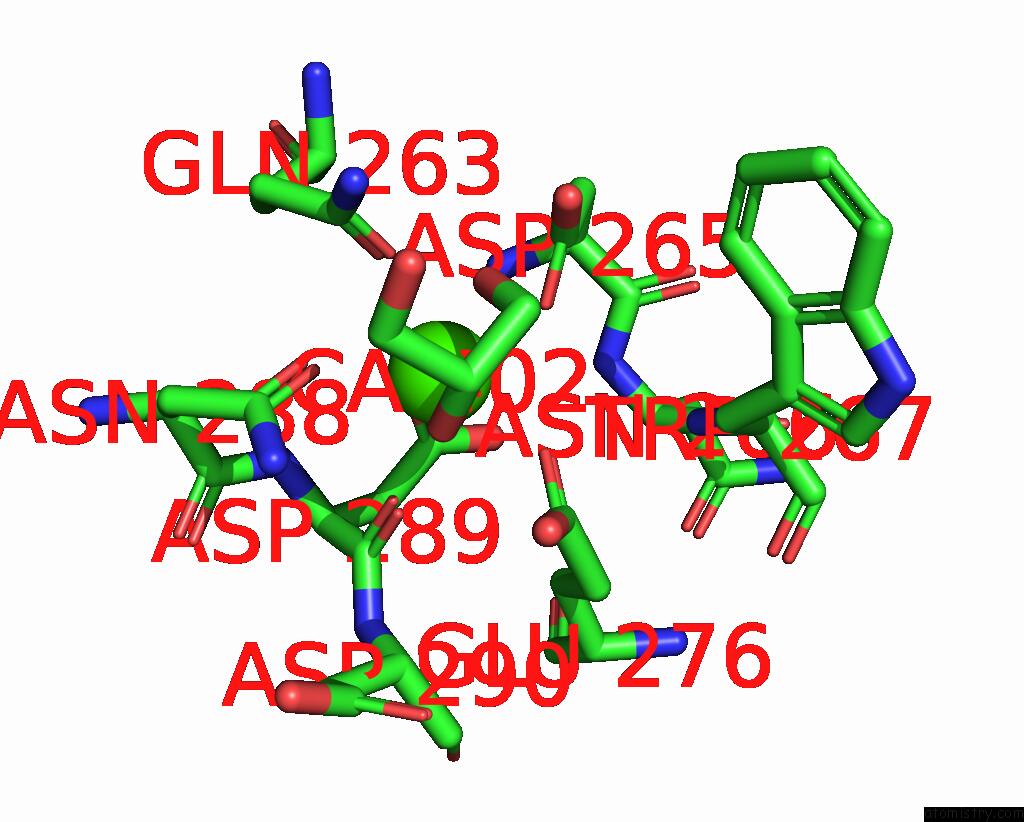
Mono view
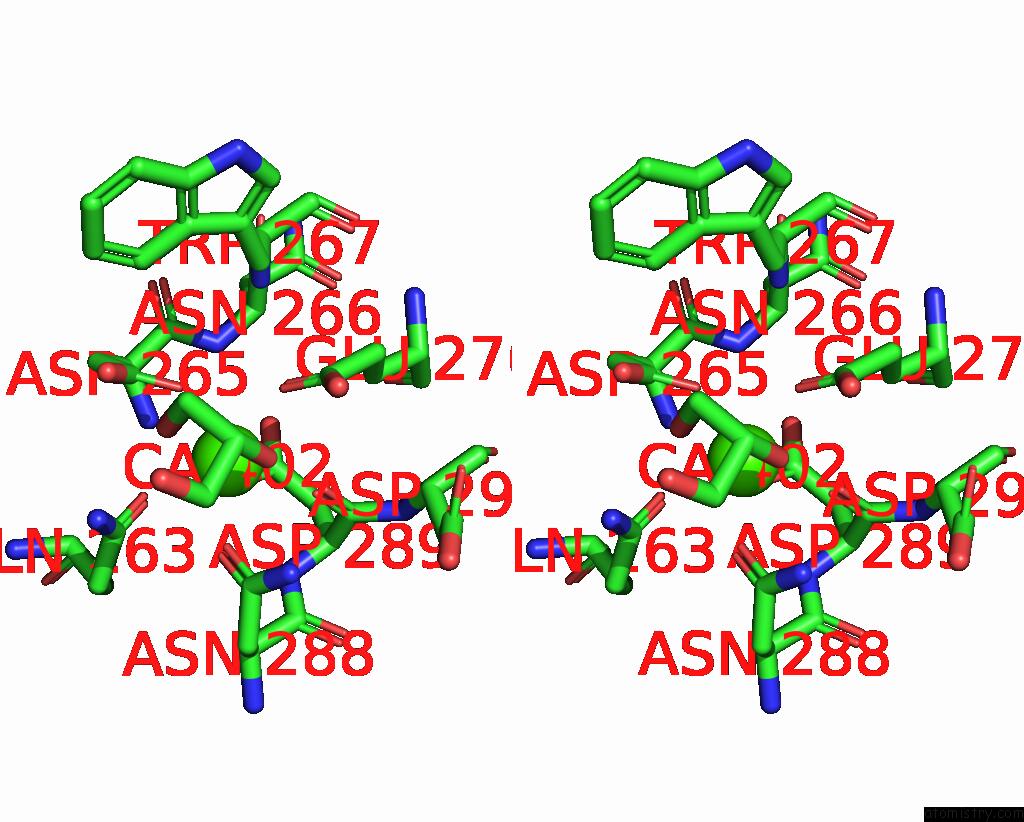
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Crystal Structure of Human ASGR2 Crd (Carbohydrate Recognition Domain) Bound to 8G8 Fab within 5.0Å range:
|
Calcium binding site 3 out of 3 in 8urf
Go back to
Calcium binding site 3 out
of 3 in the Crystal Structure of Human ASGR2 Crd (Carbohydrate Recognition Domain) Bound to 8G8 Fab
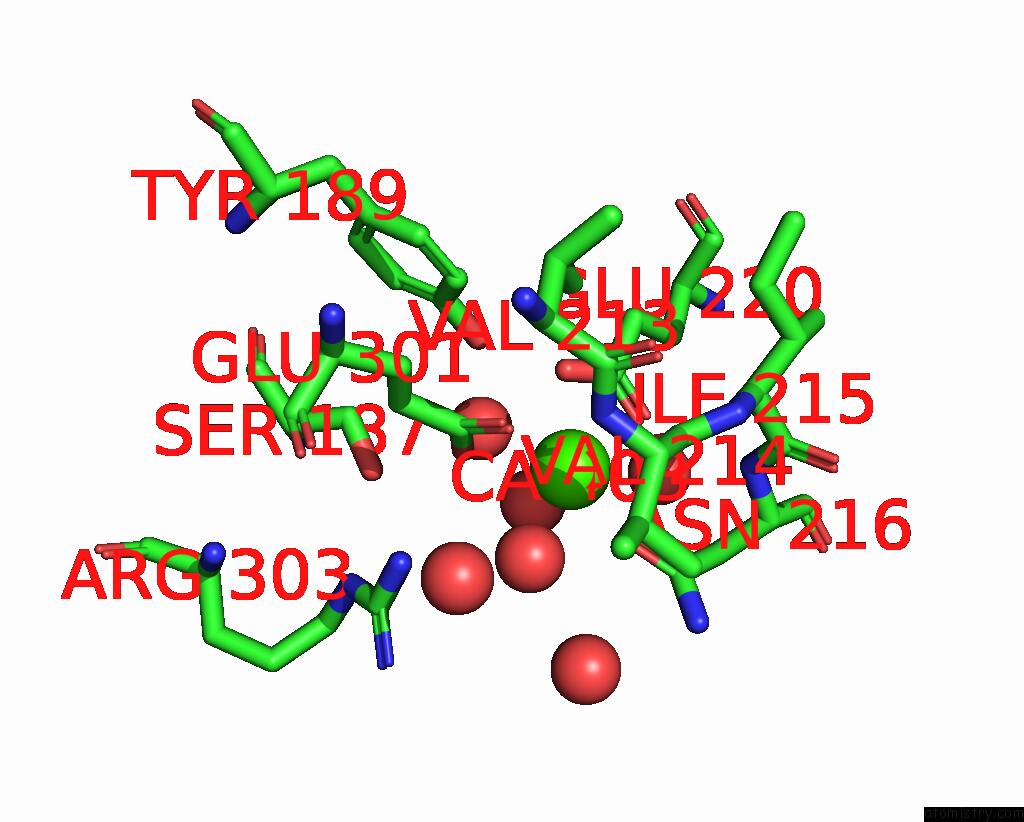
Mono view
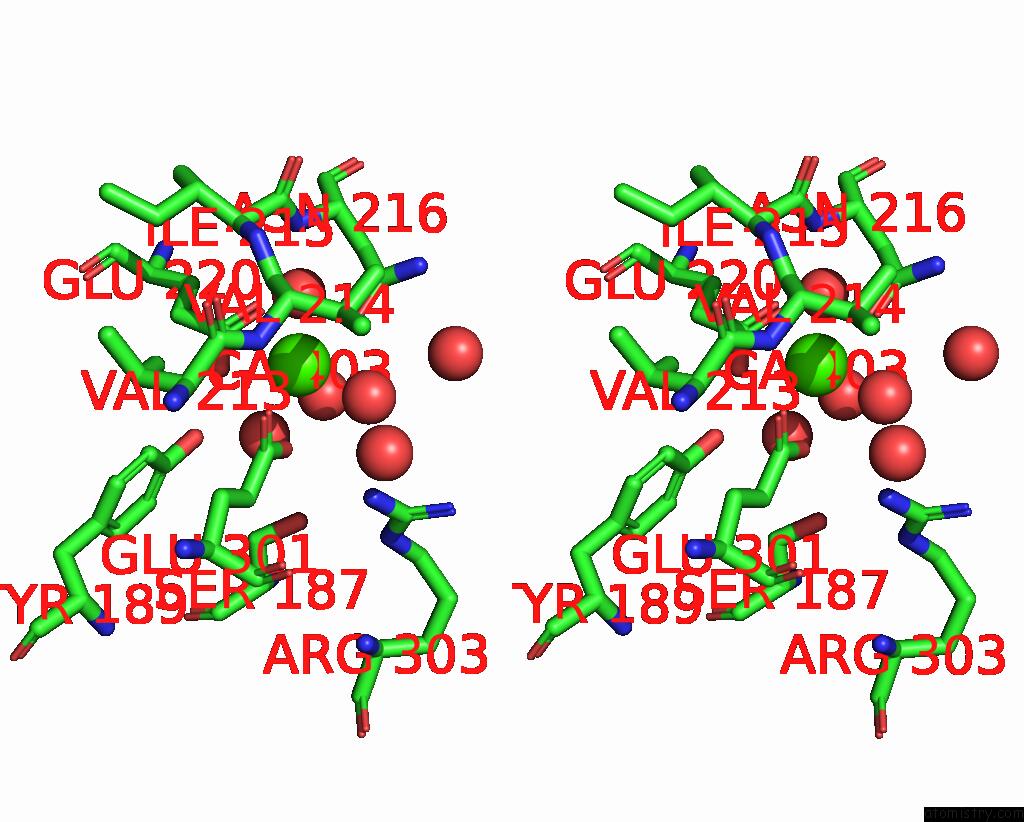
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Crystal Structure of Human ASGR2 Crd (Carbohydrate Recognition Domain) Bound to 8G8 Fab within 5.0Å range:
|
Reference:
P.Sampathkumar,
H.Jung,
H.Chen,
Z.Zhang,
N.Suen,
Y.Yang,
Z.Huang,
T.Lopez,
R.Benisch,
S.J.Lee,
J.Ye,
W.C.Yeh,
Y.Li.
Targeted Protein Degradation Systems to Enhance Wnt Signaling. Elife V. 13 2024.
ISSN: ESSN 2050-084X
PubMed: 38847394
DOI: 10.7554/ELIFE.93908
Page generated: Thu Jul 10 07:43:33 2025
ISSN: ESSN 2050-084X
PubMed: 38847394
DOI: 10.7554/ELIFE.93908
Last articles
Cl in 8CKNCl in 8CJY
Cl in 8CKP
Cl in 8CK6
Cl in 8CJF
Cl in 8CJD
Cl in 8CJO
Cl in 8CIP
Cl in 8CJ9
Cl in 8CJ8